Alene Admas1 Solomon Kiros1 Mesfin Tafesse2 Anteneh Tesfaye3 Estifanos Hawaz2 & Diriba Muleta3
11Addis Ababa Institute of Technology, Addis Ababa University, Ethiopia
2Department of Biotechnology, Addis Ababa Science and Technology University, Ethiopia
3Institute of Biotechnology, Addis Ababa University, Ethiopia
*Corresponding Author Email: aleneadmas2013@gmail.com …
Highlights
- Four yeast isolates tolerated up to 23% ethanol.
- Two isolates produced 14.3% and 13.2% ethanol from sugarcane molasses.
- Optimal production occurred at pH 4.39, 35°C, and 30° Brix.
- Isolates identified as Saccharomyces cerevisiae and Kluyveromyces marxianus.
- Results support use of indigenous yeasts for industrial bioethanol production.
Abstract
Shortage of fossil fuel supplies coupled with greenhouse gas emissions has increased interest in biofuel production and use of renewable energy worldwide. Thus, the main purpose of this study was to isolate, screen and characterize ethanol tolerant wild indigenous yeasts with high bioethanol yield from samples of local fermented alcoholic product known as areke at different stages (difdif and tinses) of production which were collected from diverse sites. The study was mainly focused to cover the following methodologies such as: isolation, screening of isolates based on the specific parameters (pH, temperature, ethanol tolerance and carbohydrate assimilation during propagation and fermentation), ethanol concentration determination and molecular identification of the potential isolates. The wild yeast isolates were retrieved from the alcoholic beverages following standard methods. From a total of 270 isolates, 10 (3.7%) of the yeast isolates tolerated 22% of ethanol. Of these 10 isolates 4 (1.5%) isolates tolerated 23% ethanol concentration, and were selected for further characterization. Ethanol production from sugar cane molasses by the yeast isolates was determined by using Hall method and was crosschecked with Ebulliometer (Bulteh 2000, India). From 4 isolates, 2 isolates (Mda and Dt1e) showed the highest bioethanol production capacity of 14.3% v/v and 13.2% v/v, respectively at pH 4.39 and temperature of 35oC in 30 degree brix molasses concentration at 150 rpm shaking condition. Based on morphological appearance of vegetative cells under microscope, colony characters and molecular analysis, the isolates were identified as Saccharomyces cereviciae, whereas one of the isolates was identified as Kluyvermyces marixianus. This study exhibited excellent bioethanol producing indigenous yeasts with high ethanol tolerance, resistant to low pH and high temperature. Therefore, isolates biomass production and utilization of the isolates for industrial bioethanol production is highly recommended.
Keywords: Areke, Difdif/Tensis, Isolation, Wild yeasts, Molasses, Fermentation, Bioethanol
1. Introduction
The energy crisis in Ethiopia prompts the study and discovery of new processes of producing renewable bioenergy such as bioethanol from sugar cane molasses using potent fermentative wild indigenous yeasts. Bioethanol is an ecofriendly and most promising biofuel that can be used in unmodified petrol engines and produced from different agro-industrial wastes such as cane or beet molasses (Lisboa et al., 2011). The combustion of bioethanol results in relatively low emission of volatile organic compounds with low evaporation, carbon monoxide and nitrogen oxides and has high biodegradable properties (Sukumaran et al., 2022). The emission and toxicity of bioethanol are lower than that of fossil fuels, such as petroleum and diesel, and this makes it the most preferred fuel in various sectors (Nwachukwu et al., 2006). Thus, there is an urgent need to produce bioethanol from different biomass sources to maximize its use in diverse sectors.
Molasses is the most preferable biomass for fermentation by yeasts to produce bioethanol because it is rich in nutrients, salts and contains easily consumable quantity of sugars, which is about 40 to 50% w/v (Demirbas, 2008). It is the final byproduct of sugar factories that is easily available and economically low in price for using it as a raw material for ethanol production (Khoja et al., 2015). In Ethiopia, currently, the only raw material for bioethanol production at the industrial level is molasses. Molasses can also be used as an alternative feed in animal fattening and also used as raw material for the production of yeast biomass and various organic acids. It is stated that from 2014 to 2020, the production of molasses increased from 0.5 million tons to 1.5 million tons and is estimated to grow to 3 million tons by 2030 (Yacob Gebreyohannes, 2013) and this sharp increment in molasses yield encourages its utilization for production of varied products including bioethanol. Generally, conversion of one ton of molasses by yeast fermentation yields 250 liters of green energy (bioethanol) (Yacob Gebreyohannes, 2013)
Therefore, bioethanol produced from renewable biomass like molasses has received considerable attention in current years. Ethanol can be produced by bacteria and yeasts (Chamnipa et al., 2017). However, yeasts have some advantages over bacteria in having efficient bioconversion ability of sugars into bioethanol with some important industrial characteristics (low nutrient requirements, ethanol resistance and tolerance to pH) (Stanley et al., 2010). Many investigations so far done have focused on the isolation and characterization of different species of wild fermentative yeasts that produce ethanol from different substrates (Wong et al., 2014). Most of these studies are focused on the isolation and characterization of wild yeast from diverse sources such as fruit (Rao et al., 2008), tej, tella and teff and molasses (Beyene et al., 2020), bagasse (Wong et al., 2014), sugar cane juices (Oliveira et al., 2008).
The Ethiopian local areke difdif contains large varieties of microbes to initiate the fermentation process and can serve as potential source of wild yeasts isolation. Areke fermentation initiation product is known as Areke-tinsis which is prepared by mixing powdered gesho leaves and powdered malt (1: 2 ratios) with water to give a mixture of free-flowing consistency and puts aside to ferment for about five days. A good amount of cereals such as finger millet (Elusine coracann) is powdered, kneaded with water to make a fermented dough. After fermentation, the dough is baked into bread. The bread is broken into pieces while warm and added to the first mixture (Yereke-tinsis) with more water and it is well mixed to make areke difdif and again left aside to ferment for about four days. Portions of the second mixture are transferred to the traditional distillation apparatus and distilled to give a product known as areke (Hotessa and Robe, 2020). Fermentation of tinsis and difdif for areke production is one of the promising sources for isolation and characterization of potential ethanol tolerant indigenous yeasts for bioethanol production (Hotessa and Robe, 2020). During ethanol fermentation, yeasts are exposed to various stresses such as ethanol they produce and which is considered to be the major stress that decreases the final yield (Nwachukwu et al., 2008). In addition, heat stress which is generated during fermentation process can greatly affect ethanol production by decreasing the growth rate of yeast strains (Techaparin et al., 2017). Ethanol production at high temperatures has gained much interest due to several advantages. For instance, a reduced risk of contaminations since ethanol-and-theromtolerant isolates can able to adapt to heat and ethanol generated during fermentation (Techaparin et al., 2017). Hence, the ability of stresses tolerance of the yeasts becomes one of the most influencing factors in ethanol fermentation using fermentative microbes (Alok et al., 2016). Even though previous research on wild yeast isolation has mostly focused on a variety of substrates, such as fruit or traditional Ethiopian beverages, there has not been any investigation into the potential applications of local Areke difdif and tensis for isolation and characterization of potential bioethanol producing native yeast strains to boost industrial-scale bioethanol production. Therefore, this study focused on the isolation and characterization of potential stress tolerant indigenous yeasts from Areke difdif and tensis.
2. Materials and Methods
2.1 Description of the Sample Collection Sites
Areke difdif and tinsis samples were collected from Debre Birhan and Dembecha districts in Amhara Regional State and from Arsi Negele district, Oromia Regional State. These sampling sites are well known for their tradition of Areke production and quality of the product in Ethiopia. All the samples were collected by using sterilized bottles and transported in ice box to Bioengineering laboratory, Addis Ababa Institute of Technology, Addis Ababa University.
2.1.1 Sampling
A total of 26 samples; 10 from Debre Berhan, 10 from Arsi Negelle and 6 from Dembecha were collected using sterile 100 ml bottles and were transported to Addis Ababa Institute of Technology, Addis Ababa University in ice box and stored at 4OC in refrigerator for further work.
2.2 Isolation of Wild Yeasts
Yeast extract peptone dextrose (YPD) agar with the composition (g/l) of Yeast Extract 10.0, Peptone 20.0, Dextrose 20.0 and Agar 15.0 was used to isolate and screen the yeasts from the collected samples. The pH of the medium was adjusted to 4.5 and after sterilization and was amended with chloramphenicol. Aliquots (0.1 ml) of appropriate separate dilutions of difdif and tinses were plated on pre-dried YPD Agar plates. All the inoculated plates were incubated for 48-72 h at 30°C (Thapa et al., 2015). Representative discrete colonies of yeast isolates were purified on the same medium and pure isolates were kept in a deep freezer with 40% glycerol for further use. For morphological analyses of the isolates, pure cultures from YPD medium were examined under a microscope (Optika, Italy).
2.3 Screening Parameters of the Yeast Isolates
2.3.1 Ethanol Tolerance
YPD liquid medium [(g/l) 6 yeast extract, 10 peptone, 6 malt extract] was used to determine ethanol tolerance of yeasts. To the cooled sterile medium, appropriate amount of absolute ethanol (99.8% v/v) was added to prepare separate duplicate tubes of 10, 15, 20, and 25% (v/v) ethanol. All the isolates were tested with 10% ethanol. The isolates that tolerated 10% were again tested at 15% ethanol and the tolerant isolates were further then tested for tolerance to 20, 22, 23, and 24% ethanol until they no more showed any growing sign. All inoculated tubes were incubated at 30°C for 24 to 48 h. The increase in optical density was recorded as evidence of growth. The concentration of alcohol at which the growth of yeasts was just inhibited was assessed as the ethanol tolerance of yeasts. The viability of each tolerant isolate was examined as indicated in Tikka et al. (2013).
2.3.2 Growth at Different pH
YPD liquid medium (broth) was used to evaluate the growth of ethanol tolerant yeast isolates in different acidic pH values. The pH of cooled sterile 150 ml of YPD broth was adjusted to 2, 2.5, 3, 3.5, and 4 in 250 ml flasks using 1M NaOH and 1M HCl. Then each flask was inoculated with half loop-full of top ten ethanol tolerant (22% and 23% v/v) yeast isolates to measure optical density at 600 nm and all the tubes were incubated at 30°C for 24 to 48 hours. Cell density was recorded at 24 and 48 h (Ghosh and Ray, 2015).
2.3.3 Growth at Different pH
Separate duplicates flasks (Ca. 150 ml) with sterile YPD broth t were inoculated with each of the ten ethanol tolerant yeast isolates and incubated at 25, 30, 35, 40, and 45°C for 48 h to observe thermo-tolerance of the selected yeast isolates. The increase in optical density of the yeast cultures at 600 nm was recorded as evidence of growth at a given temperature following standard protocol (Ghosh and Ray, 2015).
2.3.4 Carbohydrate Assimilation
The aerobic assimilation of carbon sources by the 10 ethanol-tolerant yeasts was done in sterile 20 ml of Phenol red broth (peptone 10 g, Sodium Chloride [NaCl] 5 g, Beef extract 1 g, Phenol red [7.2 ml of 0.25% phenol red solution]: 0.018 g, Carbohydrate source: 100 g [10% sugar] per liter of distilled water) in 25 ml test tube with a Durham tube. Separately inoculated tubes with the 10 ethanol-tolerant isolates in duplicates were incubated at 30°C for 24-72 h. longer incubation periods may be required to confirm a negative result. Fermentation was assessed by checking for the formation of gas (CO2) in the Durham tube and color change of the fermentation medium (deep pink to yellow) (Nasir et al., 2017). The sugars used were sucrose, glucose, lactose, maltose, xylose, and fructose).
2.4 Clarification of Molasses for Ethanol Production
Molasses was collected from Methara, Fincha, and Wonji Sugar Factories and mixed in the same ratio. In this study, 1 ml of concentrated sulfuric acid and 0.40 g urea was used in one liter (one to one diluted 500 ml molasses for clarification). Then molasses, sulfuric acid, and urea were mixed and boiled in a 1000 ml beaker at 100oC for 30 minutes. Thereafter, it was allowed to stay for 24h to cool and decant. The supernatant was used for the propagation of yeasts and fermentation to produce bioethanol (Atiyeh et al., 2003).
2.5 Molasses Preparation as a Fermentation Medium
The clarified molasses was used as a fermentation medium. In all the experiments, initial sugar concentration was measured by using Refractometer (RF-2000, UK). The fermentation medium was diluted until the molasses concentration was 40 degree brix using distilled water and autoclaved at 121° C for 15 minutes then stored as stock for propagation and fermentation (Ghosh and Ray, 2015).
2.6 Propagation of Yeasts
Separately 1 ml of 48 h old broth cultures of the ten potential yeast isolates were inoculated into each of the separate ten 500 ml flasks containing 100 ml treated and sterilized molasses. The flasks were incubated at 30oC with 150 rpm shaking condition. After 24 h, l00 ml of sterilized fresh molasses was added to each flask. The flasks were incubated for more 24 h in order to boost the cell number at 30oC. The initial (0 h) and final (48 h) ODs of the propagated cultures were measured by using UV-Vis Spectrometer (UVD-3200, LABOMED.INC, USD) at 600 nm. Finally, 200 ml molasses medium that contained 48 h propagated yeast cultures was ready as inoculum for fermentation to produce bioethanol.
2.7 Fermentation of Molasses for Ethanol Production
Submerged fermentation was carried out in 1000 ml Erlenmeyer conical flasks. Into 10 Erlenmeyer flasks of 1000 ml capacity, 300 ml of treated and sterilized molasses, and inoculum of 200 ml of propagated cells of each of the 10 yeast isolates separately were added. Flasks were cotton plugged for a partial aerobic condition for 12 h and incubated at 35oC and 150 rpm for fermentation; then the condition became anaerobic by using parafilm after 12 h and the fermentation continues for 60 h to make the total fermentation time 72 h (12 h aerobic plus 60 h anaerobic). The pH, brix of the fermentation medium, and ethanol production were recorded at 24, 48, and 72 h (Nasir et al., 2017)
2.7.1 Calculation of Ethanol Concentration by Volume
The ethanol concentration in alcohol by volume (ABV%) from the fermented medium was calculated by using a method which is based on specific gravity (SG) of the substrate as indicated in Maskell et al. (2017) by considering molasses without yeast culture as initial SG and fermented medium as final SG. The alcohol percentage of the final fermented medium would be directly proportional to the difference in the original and final (real) extract values. After all, the chemical equation for the conversion of a monosaccharide to ethanol is:
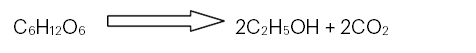
So the weight of the sugar molecule (180 atomic mass units (amu)) should be converted into the weight of two ethanol molecules (92 amu) and two carbon dioxide molecules (88 amu). This would give us an equation for the alcohol percent by weight (A%w) of:
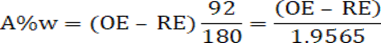
Where, OE= Original extract and RE= Real extract.
Then alcohol content as a function of the original and final specific gravities can be derived as:
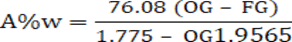
Where, OG = Original specific gravity and FG= Final specific gravity.
This equation can be converted to alcohol percent by volume (A%v) by the following formula:
A%W=A%W (FG/0.794)
Where 0.794 is the specific gravity of ethanol.
Original specific gravity and final specific gravity was measured using Digital Density Meter (DMA 4100M, UK). Ebuillometer (Bulteh 2000, India) was also used for crosschecking the alcohol concentration.
2.8 Molecular Identification of Isolates
Genomic DNA of top yeast isolates (n=10) was extracted following the modified CTAB method (Shahzadi et al., 2010). Amplification of 5.8S-ITS region of the yeast isolates was done by using ITS4 (5′-TCCTCCGCTTATTGATATGC) reverse and ITS5 (5′- GGAAGTAAAAGTCGTAACAAGG) forward universal primers in thermo-cycler, sure cycler PCR (Aglient, G8800-00, Malysia) (Gardes and Bruns, 1993). Gel purified PCR products were sequenced using Next Generation Sequencer machine illuminia, China 2010 with minor modification as per the manufacturer’s recommended protocol (Techaparin et al., 2017). The sequenced data obtained were blasted on NCBI to characterize the potent indigenous wild yeast to the species level.
The internal transcriber spacer (ITS4 and ITS5) and 5.8S of ribosomal DNA sequences were used for Basic Alignment Search Tools (BLAST). The sequence of the isolates were further aligned and compared to ITS sequences in database using the taxonomy browser of the National Center for Biotechnology Information (NCBI). A total of 20 sequences (13 from the database and 7 readable sequences from this study) were aligned using ClustalW for phylogenetic tree construction (Larkin et al., 2007). Phylogenetic tree was constructed with Molecular Evolutionary Genetic Analysis (MEGA) version 7.0 using a Maximum Likelihood (ML) algorithm character estimation method with bootstrap analysis where 1000 replicates were performed (Kumar et al., 2016).
2.9 Molecular Data Analysis
Molasses fermentation was performed in duplicate during bioethanol production and phylogenetic tree analysis was done using MEGA version 7.0 software.
3.Results
3.1 Isolation and Screening of Yeasts for Ethanol Tolerance
From samples collected, 270 yeast isolates were isolated and purified. The ethanol tolerance of the 270 purified yeast isolates is shown in Table 1. All of the yeast isolates were found tolerant to 10% and 12% but none to 24% of ethanol. The results indicated that 24 (8.9%), 10 (3.7%) and 4 (1.5%) were shown to be tolerant to 20%, 22%, and 23% of ethanol, respectively (Table 1). These isolates remained viable when grown on YEPDA plates ( Fig. 1). Those yeast isolates that tolerated 22% and 23% were considered as the best ethanol tolerant isolates and were subjected to further subsequent evaluations. The colonial feature of these isolates on YEPDA was found as white, creamy color with oval and circular shapes and had also smooth texture.
Table 1: Total number of wild yeast isolates that tolerated different ethanol concentrations
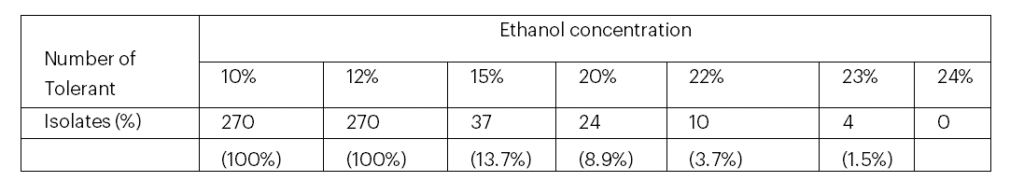
Figure 1: Growth of ethanol tolerant wild yeast isolates on solid YEPDA medium
3.2 Carbohydrates Assimilation
The results of the carbohydrate assimilation profile of the 10 ethanol tolerant (22 and 23% ethanol) yeasts are given in Table 2. Results indicated that all the 10 isolates assimilated fructose, glucose, maltose and sucrose, but only the isolate Mdb assimilated lactose and 4 isolates assimilated xylose (Table 2).
Table 2: Carbohydrate assimilation and gas production test for 10 yeast isolate
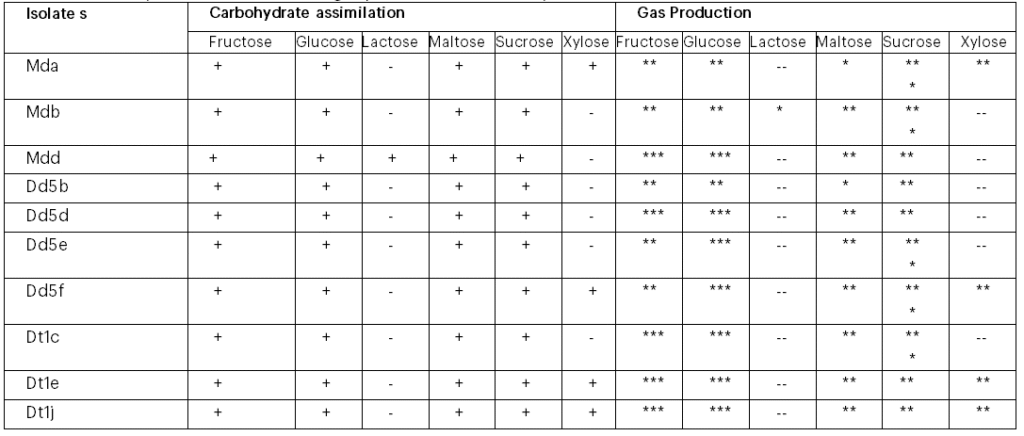
Note: *** (3) Strong gas production, ** (2) Moderate gas production, * (1) Weak gas production and (0) –/-ve no gas production. Mda=Mother difdif a, Mdb=Mother difdif b, Mdd= Mother difdif d, Dd5b= Dembecha difdif 5b, Dd5d= Dembecha difdif 5d, Dd5e= Dembecha difdif 5e, Dd5f= Dembecha difdif 5f, Dt1c= Dembecha tenses 1c, Dt1e=Dembecha tenses 1e and Dt1j= Dembecha tenses 1j.
Growth at Different Temperature and pH
The growth of the 10 ethanol tolerant yeast isolates at different temperature values (35oC, 40oC, 45oC and 47oC) is given in Table 3. All the isolates were found to be viable at 35oC, 40oC, and 45oC when allowed to grow on YEPDA plates. The isolate Mda was the only ethanol tolerant yeast that was found to be viable at 47ºC (Table 3). Ethanol-tolerant yeast isolates were able to grow at different pH values (Table 4). The results indicate that all the yeast isolates were grown at pH 2.5 and above but all failed to grow at pH 2.
Table 3: Viability of the best ethanol tolerant yeast isolates growing at different temperature values
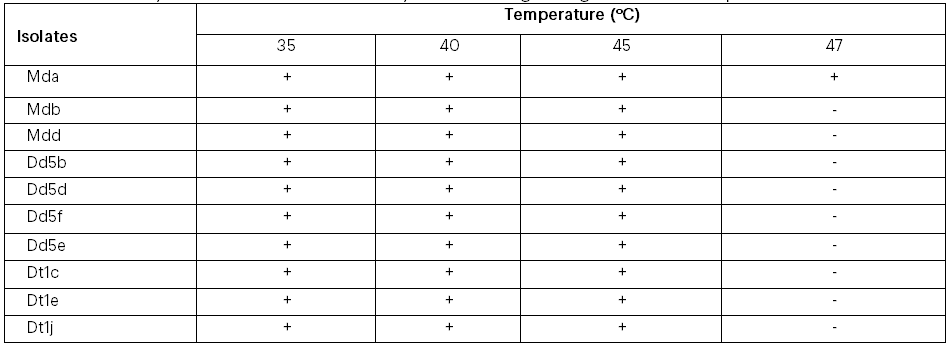
Mda=Mother difdif a, Mdb=Mother difdif b, Mdd= Mother difdif d, Dd5b= Dembecha difdif 5b, Dd5d= Dembecha difdif 5d, Dd5e= Dembecha difdif 5e, Dd5f= Dembecha difdif 5f, Dt1c= Dembecha tenses 1c, Dt1e=Dembecha tenses 1e and Dt1j= Dembecha tenses 1j.
Table 4: Tolerance of the 10 wild yeast isolates at different pH values
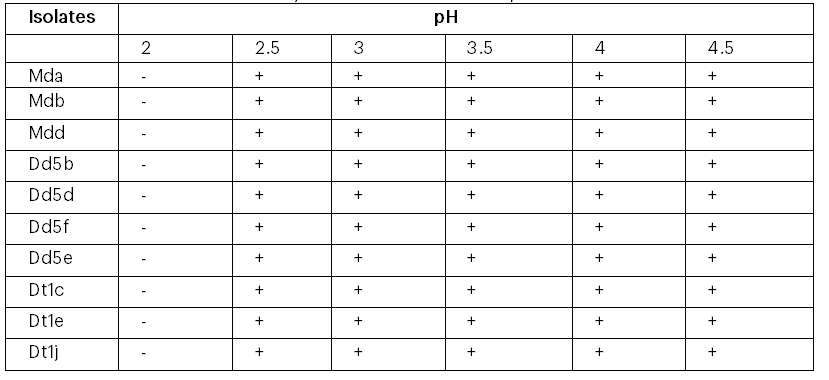
Mda= Mother difdif a, Mdb=Mother difdif b, Mdd= Mother difdif d, Dd5b= Dembecha difdif 5b, Dd5d= Dembecha difdif 5d, Dd5e= Dembecha difdif 5e, Dd5f= Dembecha difdif 5f, Dt1c= Dembecha tenses 1c, Dt1e=Dembecha tenses 1e and Dt1j= Dembecha tenses 1j.
3.3 Production of Ethanol from Molasses
The results of the potential of the ethanol production by the 10 ethanol-tolerant yeasts indicated a trend of increasing bioethanol yield by all the isolates during the course of the fermentation period (from 0 to 72 hours; Table 6). The maximum amount of ethanol (14.3% at 48 hrs and 13% at 48hrs) by volume was produced by the ethanol tolerant Mda isolate followed by the yeast isolate Dt1e (13.2%), Dt1j (12.2%), Mdd (12%), Dd5f (12%), and Dt1e (12%) at 72hrs of fermentation. The least (7.7% at 48hrs and 9% at 72hrs) ethanol was produced by isolate Dd5e (Table 6). However, better bioethanol yield was produced by the first six isolates when the fermentation time was extended to 72 hours. The pH of fermenting molasses during the production of ethanol by the yeasts remained constant, except for the yeast isolate Mda (Figure 2). The pH of fermenting molasses by AAUMda was observed rising at 48 hours (Figure 2). All the ethanol-tolerant yeast isolates exhibited the general pattern of reducing the molasses brix concentration during the course of ethanol production (Figure 3). The 4 ethanol-tolerant isolates (Mda [14.3%], Dt1e [13.2%], and Dt1j [12.2%]) were selected as the best ethanol producers at 30oB molasses and were further tested for ethanol production at 25oB and 20oB molasses.
Table 5: Mean of ethanol concentration (%V/V) produced by 10 isolates at 30 degree brix
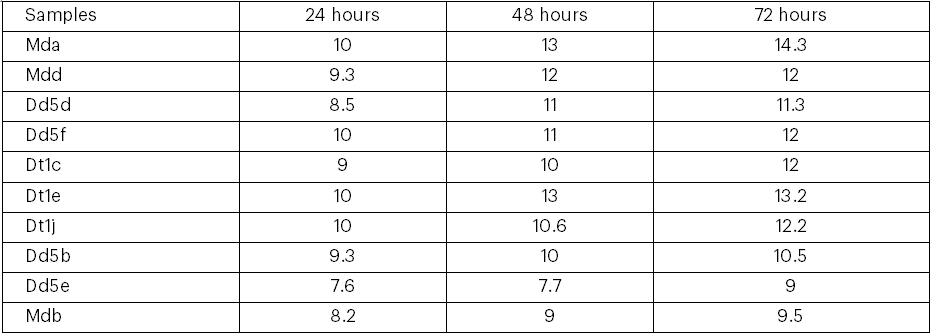
Mda=Mother difdif a, Mdb=Mother difdif b, Mdd= Mother difdif d, Dd5b= Dembecha difdif 5b, Dd5d= Dembecha difdif 5d, Dd5e= Dembecha difdif 5e, Dd5f= Dembecha difdif 5f, Dt1c= Dembecha tenses 1c, Dt1e=Dembecha tenses 1e and Dt1j= Dembecha tens es 1j.
Table 6: Summary of ANOVA and significance value between the isolates
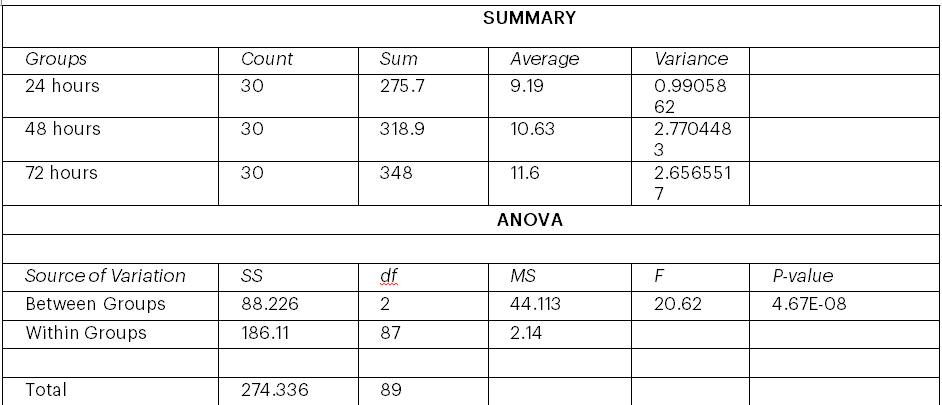
Where: SS; Sum of square, degree of freedom, MS; Mean Square, P value; significance level
This study yielded a P-value of about 4.67E-08, which indicates a very low likelihood that the observed differences could have occurred by chance alone if null hypothesis (H0) were true.
Since the P-value (<0.05) suggests that, if the null hypothesis that is, that there are no differences between the group means were true, there would be very little likelihood that the observed variations in group means are the result of pure chance. This clearly implies that at least some of the group means in your ANOVA analysis differ statistically significantly. This investigation provides strong evidence for variable levels of microbial activity or fermentation efficiency across yeast strains evaluated over a range of incubation times, which may lead to a better understanding of bioethanol synthesis.
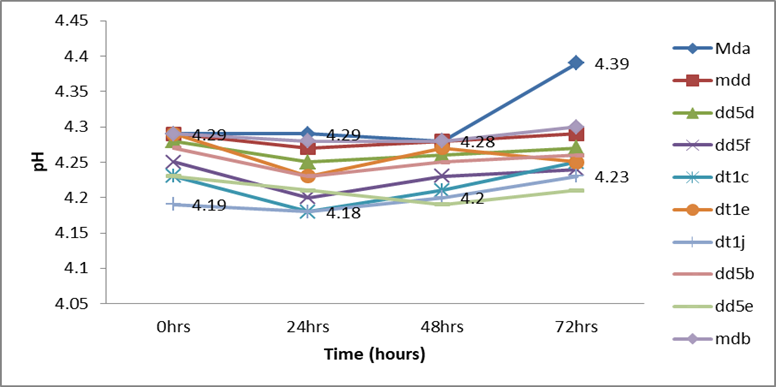
Figure 2: pH values of the fermenting medium during the production of ethanol by the ten isolates using 30°brix molasses concentration at 35oC.
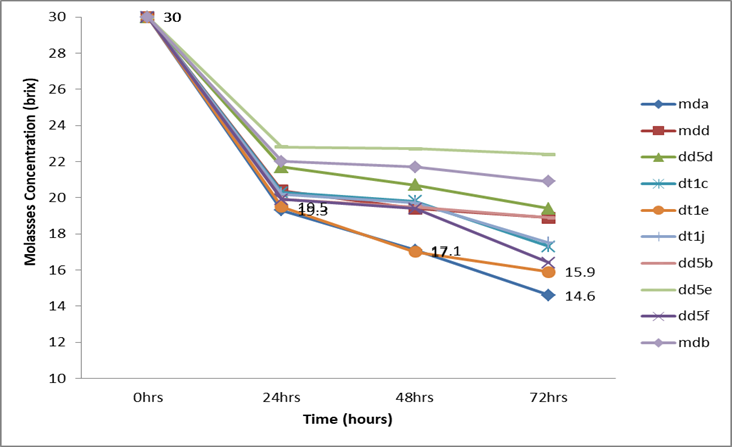
Figure 3: Sugar brix change for the ten wild yeast isolates during ethanol production from molasses.
The results of the best 4 ethanol producers at 20oB indicated that the highest ethanol yield was recorded by the isolate Mda (7.5%) and Dt1e (7.1%) at 48 hours and Dt1j (8.1%) and Mda (7.8%) at 72 hours, whereas the lowest yield (6.8%) at 48 hours was recorded by isolate Dd5f and Dt1j (Figure 3, 4 and 5). All the four isolates consumed above 48% sugar concentration at 48 hours and 50% at 72 hours fermentation but the ethanol yield was very low compared to their production at 30oB with reduced pH values from 5.6 to 4.73 in 72 hours (Figure 3, 4 and 5).
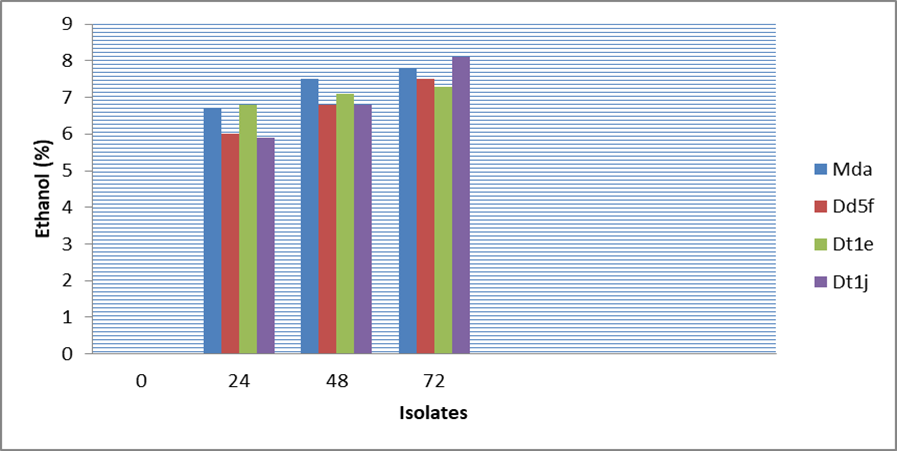
Figure 4: Ethanol production ability of 4 isolates from 20º brix molasses concentration at 35oC.
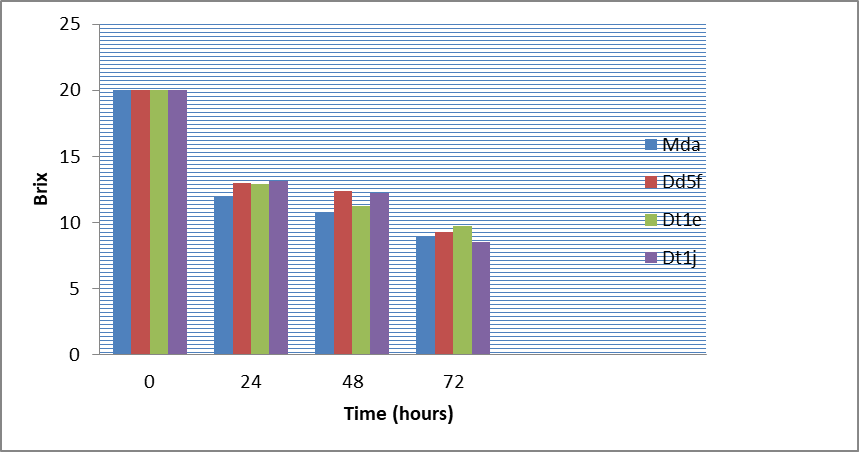
Figure 5: Sugar brix (20oB) dynamics for 4 top isolates as fermentation time increases
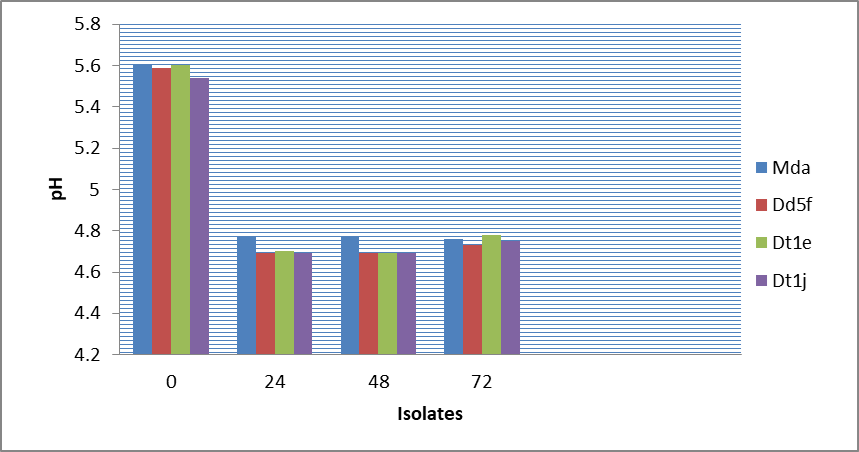
Figure 6: Change in pH values during the production of ethanol by the 4 isolates using 20 brix molasses concentration at 35oC.
With 25oB molasses concentration, the ethanol yield recorded was almost the same (11%) for all the four isolates up to 72 hours, except slightly lower yield at 24 hours (Figure 7). The sugar brix was reduced from 25oB to 9oB again and almost the same for all the four isolates at 72 hours (Figure 7) with pH reduction from 5.6 to 4.85 for all the four isolates in 72 hours (Figure 8). The comparative results of ethanol production by the 4 best yeast isolates showed that the highest ethanol production was recorded from 30oB in 72 hours (Figure 8), whereas the lowest ethanol was recorded from 20oB at 72 hours
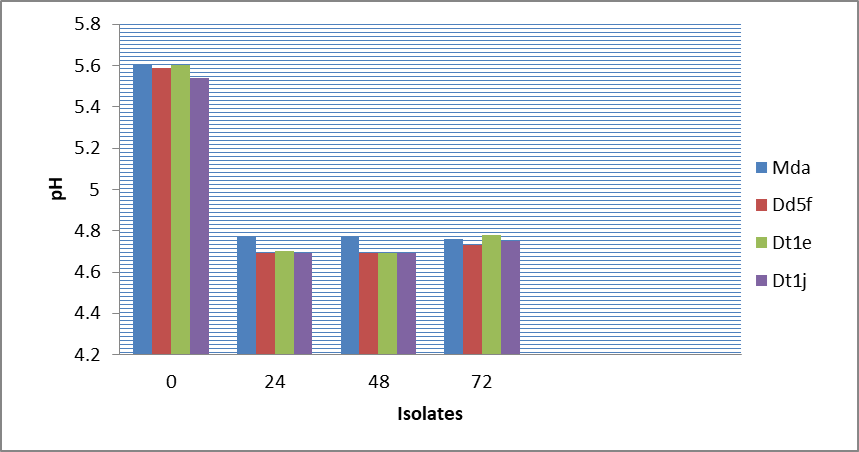
Figure 7: Ethanol production of 4 isolates from 25 brix molasses concentration
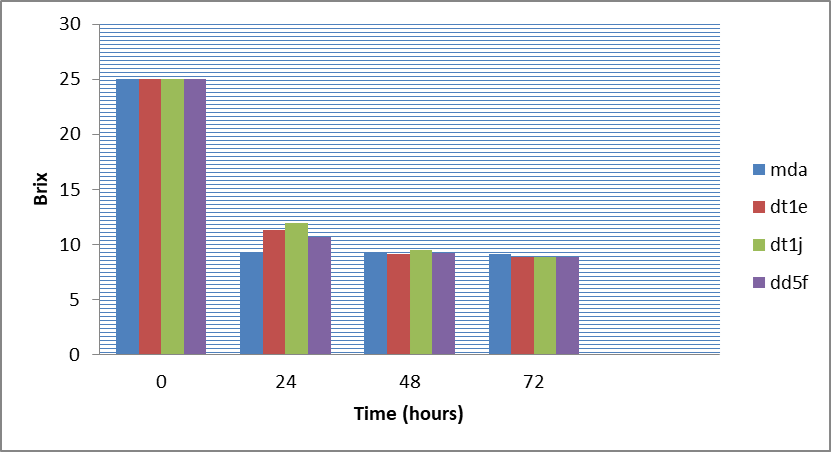
Figure 8: Change in brix (25ºB ) as time increases
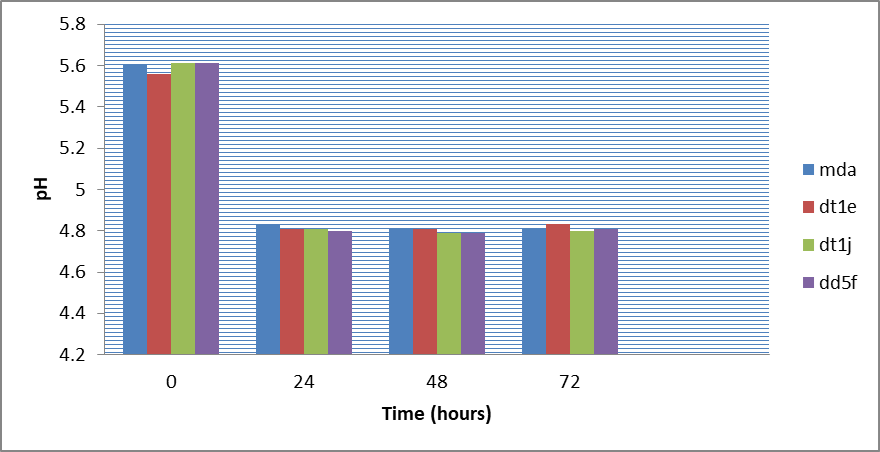
Figure 9: Change in pH values during the production of ethanol by the 4 top isolates using 25 brix molasses concentration at 35oC
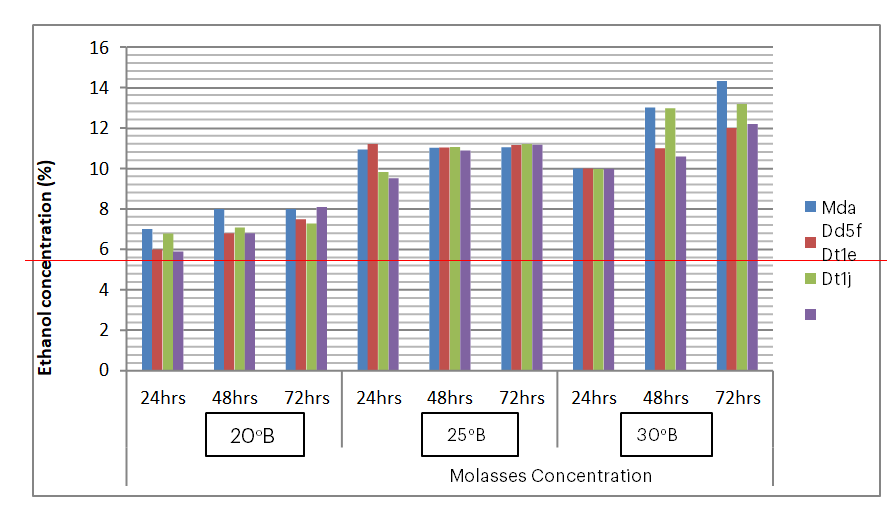
Figure 10: Ethanol yield from top 4 fermenters with different molasses concentration at 24, 48 and 72 hours
3.4 Molecular Analysis
The results of the DNA extracts, PCR amplifications, BLAST search and their phylogenetic tree for DNA sequences of the best ethanol tolerant yeasts are shown in Figure 10, Table 7 and Figure 11 in that order. All the amplified PCR products of the isolates were between 830 to 850 bp lengths (2.5 kb) (Figure 10). According to the BLAST result from the ten isolates, only seven isolates showed similarity (92-99%) to gene bank data entries, whereas the three isolates did not match with any of yeast strains in the Gene Bank. The highest similarity (99%) was shown between Mdb and Saccharomyces cerevisiae E2598a and AAUDd5b and Saccharomyces cerevisiae strain UOA/HCPF5911. Besides, the results of this study indicate that isolate Mdd is 98% similar to Kluveromyces marixianus MOA, whereas isolates Mda, Dd5f, d5e and Dd5b are 94, 96, 98 and 92% similar to S. cerevisiae strain GP2, S. cerevisiae isolate E21567, S. cerevisiae strain UCDFST11-194 and S. cervisiae NGL3A, respectively (Table 7). In this study, the isolates were belonged to two genera Saccharomyces and Kluveromyces. However, only the isolate Mdd was assigned to genus Kluveromyces and the rest six isolates were included under Saccharomyces genus.
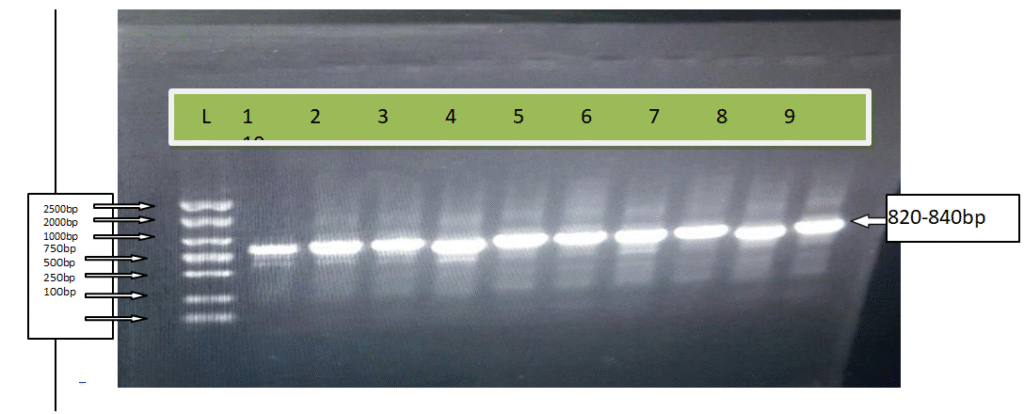
Figure 11: Products of PCR for ten yeast isolates using primers ITS4 and ITS5. Where; L: Ladder, 1:Dt1e, 2:Dd5d, 3:Dd5b, 4:Dt1j, 5: Mdd, 6:Dd5f, 7: Mdb, 8: Mda, 9:Dt1c and 10:Dd5e
Table 7: BLAST similarities of the sequenced seven isolates from NCBI database
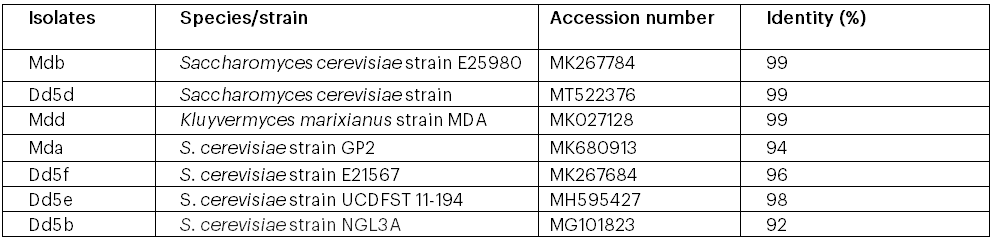
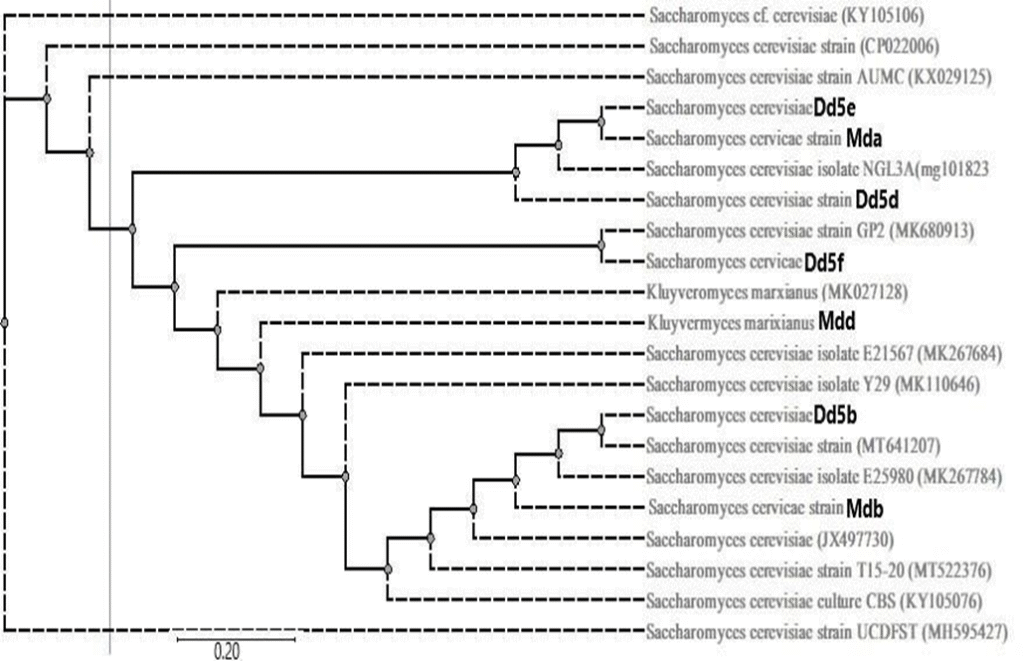
Figure 11: Phylogenetic tree analysis of seven wild yeast isolates from areke difdif/Tensis using ITS DNA sequence.
Seven isolates Mda, Mdb, Mdd, Dd5f, Dd5e, Dd5d, and Dd5b with their accession numbers and the 14 similar strains from gene bank was (MG101823, KY105106, CP022006, KX029125, MK680913, MK027128, MK267784, MK267684, MH595427, MK110646, MT641207, JX497730, KY105076 and MT522376).
4. Discussion
In this study a total of 270 isolates were isolated and characterized. Morphological characterization of all isolates using colonial and microscopic observation seemed to indicate that all of the isolates were yeasts that are members of Saccharomyces species. Microscopic observation further confirmed oval and circular shaped individual cells that reproduced by budding. Similar conclusion was reached on the basis of growth pattern studies in liquid and solid YPD media; in all cases white and creamy colonies with smooth colony texture were obtained after subculture by Thapa et al., (2015).
In this study top ethanol tolerant yeast isolates were screened based on their tolerance to different concentration of ethanol. All isolates (270) were found tolerant to 10% and 12% ethanol concentration; but 37, 25, 10 and 4 isolates were found tolerant to 15%, 20%, 22% and 23% ethanol concentration, respectively (Table 1). Ethanol is the main inhibitor of yeast growth at relatively low concentrations by inhibiting cell division; decreasing cell biomass and specific growth rate; in addition to that high ethanol concentrations, reduces cell vitality and increases cell death also it influences cell metabolism and macromolecular biosynthesis (Stanley et al., 2010 ; Hu et al., 2012). According to Techaparin et al. (2017) specified the percentage of survivals of thermotolerant yeast isolates in YM medium at 7, 10 and 13% ethanol concentration.
Saccharomyces cervisiae isolated from Raffia palm wine (TBY 1 and TGY 2) exhibited resistance to 15.5% and 16% v/v ethanol concentration, respectively (Nwachukwu et al., 2006). Almost similar tolerance at 16.5% v/v ethanol has been observed for isolate HJ33 followed by the 16% v/v ethanol tolerant yeast isolate HJ12 in the broth medium (Iticha, 2016). Sacchromyces cervisiae of palm wine origin by protoplast fusion showed the highest tolerance of 20% v/v ethanol (Nwachukwu et al., 2008). In present study ten isolates, were observed tolerating more than 20% ethanol. Interestingly, isolates Mdb, Mda, Dd5d and Dd5f were found tolerant to 23% ethanol concentration and the other 6 isolates were found tolerant to 22% ethanol concentration.
In this study the carbohydrate assimilation test was carried out for 10 ethanol tolerant isolates. All 10 isolates in this study showed consumption of the six carbon sugars included in the study. This could be an important indication of isolates to convert the given sugars in to the needed byproducts like ethanol. One isolate, Mdd was shown utilizing lactose and four isolates, Mda, Dd5f, Dt1e and Dt1j were observed utilizing the five carbon sugar, xylose (Table 2). Yeasts especially members of Saccharomyces species were shown utilizing glucose, fructose and sucrose and Pichia stiptis can able to assimilate 5 carbon sugars (xylose) (Tsegaye et al., 2021). These indicate that the isolates found in this study are broad sugar fermenters and showed their potential of bioethanol productivity.
Thermo tolerance test also specified that all the ten isolates of this study grew best at 30, 35, 40, and 45oC within 24 hours of incubation period in YPD solid and liquid medium. It was found that isolate Mda of this study was able to grow at 47oC (Table 3). The optimum growth temperature for members of Saccharomyces species was observed at 30oC and optimum temperature for higher ethanol production was 35oC indicated by Sukumaran et al. (2022). According to Ali and Khan, (2014) strains C2 and TA were shown growing well up to 37oC, but showed weak growth at 40oC and inadequate growth at 42oC. The finding in this study has indicated that the isolates were tolerant to high temperature; which makes them industrially useful and promising to survive sustainably at temperature stress environment for better ethanol production. It was indicated that isolate Mda was able to grow at 47oC unlike the result given by (Ali and Khan, 2014). According to Chamnipa et al. (2018) 62 isolates those were obtained from soil and rotten fruits were shown growing at 37oC, whereas 40 isolates were grown at 40 and 45oC. Therefore, all the ten isolates from this study could be classified as thermo tolerant yeasts since they were able to grow at 40, 45 and one isolate at 47oC. If yeast can able to grow above maximum temperature range of mesophilic yeast, from 30 to 40oC it, is considered to be thermo tolerant yeasts (Sree et al., 2000).
Growth at different pH values of the entire selected top 4 isolates showed growth at pH 2.5, 3, 3.5, 4 and 4.5. However, all were not able to grow at pH 2 (Table 4). According to Sukumaran et al. (2022) the isolates that were grown at 2 to 5 pH range produced maximum ethanol at pH 4. In this study it was found that the isolate Mda showed maximum ethanol production at pH 4.4.
The amount of bioethanol production by the 4 ethanol-tolerant yeast isolates was examined at 35oC, shaking at 150 rpm, and pH adjusted to 4 to 5.0. In the present study, the amount of bioethanol production of the four isolates from molasses, with brix of 20, 25, and 30 degree brix (oB) ranged from 8 to 14.3% (v/v) of ethanol yield. Isolate Mda which was able to tolerate high ethanol concentration (23%), produced the highest amount of ethanol yield (14.3% v/v) with 30oB at 35oC, whereas, the lowest ethanol yield was obtained from isolate Dd5e (9% v/v) at the same temperature and sugar concentration with Mda isolate. Ethanol concentration produced from alcohol yeast and New Aule Baker’s Yeast were 7.48% and 10.29% v/v without initial aeration and 8.7% and 12% v/v with the initial aeration at 38oC, respectively (Mayzuhroh et al., 2016). Aeration in the early stage of fermentation is important for yeast to synthesize cell membrane, especially sterols and unsaturated fatty acids which are essential to assure cell membrane integrity and to vent CO2 that inhibits yeast cell growth Khongsay et al., 2012). This result was found similar to isolates in this study that produced the average maximum of ethanol yield that ranged from 11% to 14.3% v/v with early stage aeration at 35oC. The aeration of 0.2 vvm under fed-batch fermentation increased 23% of cell viability (Aldiguier et al., 2004). On the average, ethanol yields from sugarcane molasses were found to be within the range of 7.8% v/v in another similar study done by (Nofemele et al., 2012). In line to this study, based on molasses concentration at 72 hours, the highest ethanol yield was obtained from 30oB by isolate Mda with the yield of 14.3% v/v, whereas the lowest ethanol yield was obtained from 20oB by isolate Dt1e with the yield of 7.3% v/v. Once more, at 24 hours and 48 hours, the highest ethanol yield obtained from 30oB molasses concentration was 10% and 13% from isolate Mda (Table 5). Thus, this actually indicates that the selected isolates could produce the lowest ethanol yield from 20 and 25oB molasses concentration (Figure 4 and 7). The mutant yeast (YM4) created by applying 266 nm radiation produced 11.9% v/v ethanol at 35oC in 72 hours with 100 rpm shaking condition (Zhang et al., 2014). This result significantly differ from result of this study in which the isolates were able to produce above 12% v/v ethanol without any attempt of strain improvement. According to Ethiopian sugar factory data, the commercial industrial ethanol producing yeast in fermentation tank be able to produce 8- 9% v/v ethanol at 32-35oC with pH of 4.8 to 5 in 48 hours from 25-30 brix molasses concentration (Ethiopian sugar factory, 2020). So the isolates in this study showed better performance in ethanol tolerance and production than that of the commercial industrial yeasts used by sugar factories at Fincha and Metehara ethanol plants.
The highest sugar consumption was observed by isolate Mda at 72 hours which consumed more than 50% of sugar and reduced the brix from 30 to 14.6oB; high sugar consumption by this isolate in an important indication of the potential of producing high ethanol yield. Whereas the lowest sugar consumption was observed by isolate Dd5d which consumed 35% of sugar with the brix reduction from 30 to 19.4 at 72 hours as indicted in Figure 3, due to lower sugar consumption, this isolate showed low ethanol production comparing to the other isolates.
Molecular level identification of the ten isolates in this study was done and all isolates were subjected to total genomic DNA extraction and the internal transcriber spacer (ITS) region for each isolates was amplified using ITS 4 (5′-TCCTCCGCTTATTGATATGC) and ITS 5 (5′-GGAAGTAAAAGTCGTAACAAGG) primers giving 820-840 bp of PCR product (Figure 10). The amplified PCR result from each isolate was sequenced and the sequences were analyzed using BLAST at NCBI. Isolate Mdb and Ddfd showed 99% similarity to S. cervisiae strain E25980 and S. cervisiae strain UOA/HCPF5911, respectively from the NCBI data base; whereas isolate Dd5f and Dd5e showed 96% and 98% similarity to S.cervisiae strain E21567 and S.cervisiae strain UCDFST 11-194 respectively; however, isolate Mdd showed 98% similarity to Kluyvermyces marixianus isolate MDA (Table 7). This similarity values indicted that it was above the threshold for genus and species identification. Isolate Mda and Dd5b showed 94% and 92% similarity to S. cervisiae strain GP2 and S. cervisiae isolate NGL3A respectively (Table 7), however, this similarity values are below the threshold for genus and species identification, consequently they could be a new genus or species (Oslan et al., 2012). The other three isolates (Dt1c, Dt1e and Dt1j) were did not show any similarities from the NCBI database.
Phylogenetic study for the isolates was plotted and the cladogram tree obtained by using MEGA version 7 Tamura et al. (2007) was shown in Figure11. To evaluate the reliability of the inferred tree the option of doing bootstrap analysis was followed as indicated in (Oslan et al., 2012). The bootstrap value is attached to each branch and this value is a measure of confidence in this branch, so the maximum value was 100 and minimum value was 55 with 0.02 tree distance matrix (Figure 11).
Many wild type yeast strains were subjected to mutagenesis process to create mutant yeasts for better industrial performance. This approach is costly and time consuming, so it is very vital to isolate and characterize efficient, stress tolerant and effective industrially persuasive indigenous yeasts from local and natural sources (Zhang et al., 2012). This study managed to achieve its objective, in contrast to the above studies and by isolating stress tolerant strains from local sources for industrial scale bioethanol production. Physiological studies on the ten isolates and molecular tests on seven isolates yielded the conclusion that isolates were strains of S. cervisiae except the isolate Mdd which was found to be member of Kluyveromyces spp.
5. Conclusion
The present study revealed that the ten isolates showed excellent performance in ethanol tolerance and temperature tolerance as well; they were also found surviving at various pH ranges. The ability to produce ethanol from sugarcane molasses was tested for all top ten stress-tolerant isolates. The identified stress tolerant yeast isolates (Mda, Dt1e, Dt1j, and Dd5f) have a high resistance to ethanol and can flourish in a different ranges of pH and temperature. This remarkable endurance enables more effective fermentation operations, which can result in increased bioethanol production yields, particularly in demanding industrial environments. The study’s emphasis on turning sugarcane molasses into bioethanol showed how to produce biofuel in a sustainable manner. In addition to giving waste materials a valuable new purpose, using agricultural and industrial by-products lowers greenhouse gas emissions and trash disposal problems that are usually brought on by their breakdown.
Increased ethanol production efficiency could result in reduced production costs, which would make bioethanol a more attractive substitute for fossil fuels. This can promote economic expansion, especially in sugarcane-producing nations, and help create jobs in the biofuel industry. The successful fermentation of molasses to produce ethanol can lead to lower carbon footprints if widely adopted, replacing fossil fuel dependency with renewable energy sources. This transition is critical to combat climate change and promote cleaner energy solutions.
6. Recommendations
The following recommendations must be taken into account in order to expand on these discoveries and optimize the potential of the identified yeast isolates for increasing the production of bioethanol:
- Perform genomic and proteomic studies on the high-performing isolates to identify key genes and proteins involved in stress tolerance and ethanol production. This foundational knowledge can guide targeted biotechnological improvements.
- To verify the top four isolates’ ability to function in higher volumes, do pilot-scale fermentation experiments with them in various settings. To go from lab-scale results to commercial feasibility, this stage is crucial.
- Examine the yeast strains’ metabolic processes to gain a better understanding of their tolerance mechanisms and capacity to produce ethanol. This information can help with metabolic or genetic engineering to improve their performance even more.
- Encourage collaborations with the agricultural and biofuel industries to test these discoveries in practical contexts, promoting knowledge sharing and the commercialization of the study’s findings.
Acknowledgement
The author is grateful to the Bioengineering Laboratory, School of Chemical and Bio-Engineering, Addis Ababa Institute of Technology, Addis Ababa University, Ethiopia for supporting the equipment and facilities for this research. Also the author is grateful to the bioethanol research team.
Conflicts of Interest
The authors declare that they have no competing interests regarding the publication of this paper.
Authors’ Contributions
Alene Admas, Solomon Kiros, Mesfin Tafesse, Anteneh Tesfaye, Estifanos Hawaz and Diriba Muleta did the proposal write up and sample collection. Alene Admas, did isolation and characterization, fermentation dynamics data analysis, and manuscript preparation. Alene Admas, did molecular identification of the yeast isolates. Alene Admas did sequence data analysis. Diriba Muleta, Anteneh Tesfaye, Solomon Kiros, did manuscript edition. The author(s) read and approved the final manuscript.
References
Aldiguier, A. S., Alfenore, S., Cameleyre, X., Goma, G., Uribelarrea, J. L., Guillouet, S. E., & Molina-Jouve, C. (2004). Synergistic temperature and ethanol effect on Saccharomyces cerevisiae dynamic behaviour in ethanol bio-fuel production. Bioprocess and biosystems engineering, 26(4), 217-222.
Ali, M. N., & Khan, M. M. (2014). Screening, identification and characterization of alcohol tolerant potential bioethanol producing yeasts. Curr Res Microbiol Biotechnol, 2(1), 316-324.
Alok, J., Tomer, D. and Tripti, B (2016). Microbial and Biochemical Technology Bioethanol Production by Novel Indigenous Yeast Strains from Lignocellulosic Waste. 8(6):474-477.
Atiyeh, H. and Duvnjak, Z (2003). Production of fructose and ethanol from cane molasses using Saccharomyces cerevisiae ATCC 36858. Acta Biotechnol, 23(1):37-48.
Beyene, E., Tefera, A. T., Muleta, D., Fantahun, S. K., & Wessel, G. M. (2020). Molecular identification and performance evaluation of wild yeasts from different Ethiopian fermented products. Journal of Food Science and Technology, 57, 3436-3444.
Chamnipa N., Thanonkeo S., Klanrit, P (2017). Biotechnology and Industrial Microbiology The potential of the newly isolated thermotolerant yeast Pichia kudriavzevii RZ8-1 for high-temperature ethanol production. Brazilian J Microbiol, 49(2):378-391.
Chamnipa, N., Thanonkeo, S., Klanrit, P., and Thanonkeo, P. (2018). The potential of the newly isolated thermotolerant yeast Pichia kudriavzevii RZ8-1 for high-temperature ethanol production. Brazilian journal of microbiology, 49, 378-391.
Demirbas, A. (2008). Biofuels sources, biofuel policy, biofuel economy and global biofuel projections. Energy conversion and management, 49(8), 2106-2116.5.
Gardes M. and Bruns, T. D (1993). ITS primers with enhanced specificity for basidiomycetes ‐ application to the identification of mycorrhizae and rusts. Mol Ecol., 2(2):113-118. 25.
Ghosh, D. and Ray, D. (2015). Effects of pH on growth of Yeast isolates in different media. Journal of Mycology, 1(2):157-167
Hotessa, N., & Robe, J. (2020). Ethiopian indigenous traditional fermented beverage: the role of the microorganisms toward nutritional and safety value of fermented beverage. International Journal of Microbiology, 2020(1), 8891259.
Hu, N., Yuan, B., Sun, J., Wang, S. A., & Li, F. L. (2012). Thermotolerant Kluyveromyces marxianus and Saccharomyces cerevisiae strains representing potentials for bioethanol production from Jerusalem artichoke by consolidated bioprocessing. Applied microbiology and biotechnology, 95, 1359-1368.
Iticha, T. N. (2016). Isolation and screening of ethanol tolerant yeast for bioethanol production in Ethiopia. Global Journal of Life Sciences and Biological Research, 2(2), 1-7.
Khoja, A. H, Ali, E, Zafar K., Ansari, A. A and Nawar, A (2015). Comparative study of bioethanol production from sugarcane molasses by using Zymomonas mobilis and Saccharomyces cerevisiae. 14(31):2455-2462.
Khongsay, N., Laopaiboon, L., Jaisil, P., & Laopaiboon, P. (2012). Optimization of agitation and aeration for very high gravity ethanol fermentation from sweet sorghum juice by Saccharomyces cerevisiae using an orthogonal array design. Energies, 5(3), 561-576.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution, 33(7), 1870-1874.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H. and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0. bioinformatics, 23(21), 2947-2948.
Lisboa, C. C., Butterbach‐Bahl, K. L. A. U. S., Mauder, M., & Kiese, R. (2011). Bioethanol production from sugarcane and emissions of greenhouse gases–known and unknowns. Gcb Bioenergy, 3(4), 277-292.
Maskell, P. D., Alex Speers, R. and Maskell, D. L (2017). Improving uncertainty in Widmark equation calculations: Alcohol volume, strength and density. Sci Justice, 57(5):321-330.
Mayzuhroh, A., Arindhani, S., & Caroenchai, C. (2016). Studies on bioethanol production of commercial baker’s and alcohol yeast under aerated culture using sugarcane molasses as the media. Agriculture and Agricultural Science Procedia, 9, 493-499.
Nasir, A., Rahman, S. S., Hossain, M. M. and Choudhury N (2017). Isolation of Saccharomyces cerevisiae from pineapple and orange and study of metal’s effectiveness on ethanol production. Eur J Microbiol Immunol., 7(1):76-91.
Nofemele, Z., Shukla, P., Trussler, A., Permaul, K., & Singh, S. (2012). Improvement of ethanol production from sugarcane molasses through enhanced nutrient supplementation using Saccharomyces cerevisiae. Journal of Brewing and Distilling, 3(2):29-35.
Nwachukwu, I. N, Ibekwe, V. I, Nwabueze, R. N, Anyanwu, B. N (2006). Characterisation of palm wine yeast isolates for industrial utilisation. African J Biotechnol, 5(19):1725-1728.
Nwachukwu, I. N., Ibekwe, V. I. and Nwabueze, R. N (2008). Production of high-ethanol-yielding Saccharomyces cerevisiae of palm wine origin by protoplast fusion. Journal of Life science, 5(4):64-68.
Oliveira, A., Arau, M. and Fietto, L. G (2008). Biochemical and Molecular Characterization of Saccharomyces cerevisiae Strains Obtained from Sugar-Cane Juice Fermentations and Their Impact in Cachac. 74(3):693-701.
Oslan, S., Salleh, A., Rahman, R., Basri, M., & Chor, A. (2012). Locally isolated yeasts from Malaysia: Identification, phylogenetic study and characterization. Acta Biochimica Polonica, 59(2), 225-229.
Rao, R. S., Bhadra, B., & Shivaji, S. (2008). Isolation and characterization of ethanol‐producing yeasts from fruits and tree barks. Letters in applied microbiology, 47(1), 19-24.
Shahzadi, I., Ahmed, R., Hassan, A., and Shah, M. M (2010). Optimization of DNA extraction from seeds and fresh leaf tissues of wild marigold (Tagetes minuta) for polymerase chain reaction analysis. Genet Mol Res., 9(1):386-393.
Sree, N. K., Sridhar, M., Suresh, K., Banat, I. M., and Rao, L. V. (2000). Isolation of thermotolerant, osmotolerant, flocculating Saccharomyces cerevisiae for ethanol production. Bioresource Technology, 72(1), 43-46.
Stanley, D., Bandara, A., Fraser S., Chambers, P. J and Stanley, G. A (2010). The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. 109:13-24.
Sukumaran, R. K., Christopher, M., Sreeja-Raju, A., Sankar, M., Kooloth-Valappil, P., Gnanaraj, V. R. and Adarsh, V. P. P. (2022). Utilization of Agro-Industrial Wastes for Biofuel Generation. In Valorization of Agro-Industrial Byproducts (pp. 285-314). CRC Press.
Tamura, K., Dudley, J., Nei, M., & Kumar, S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular biology and evolution, 24(8), 1596-1599.
Techaparin, A., Thanonkeo, P. and Klanrit, P (2017). High-temperature ethanol production using thermotolerant yeast newly isolated from Greater Mekong Subregion. Brazilian Journal of Microbiology, 48(3):461-475.
Thapa, S., Shrestha, R., Tirewal, A. and Sharma, A. K. C (2015). Isolation of yeast from soil and different food samples and its characterization based on fermentation. Nepal Journal Biotechnology, 3(1):29-34.
Tikka, C., Osuru, H. P., Atluri, N., Raghavulu, P. C. V., Mannur, I. S., Prasad, U. V. and Bhaskar, M. (2013). Isolation and characterization of ethanol tolerant yeast strains. Bioinformation, 9(8), 421.
Tsegaye, M., Chandravanshi, B., Feleke, S., & Redi-Abshiro, M. (2021). Enhanced cellulose efficiency of pressurized hot water pretreated highland Ethiopian bamboo (Yushania alpina): A potential feedstock for ethanol production. Chemistry International, 7(1), 53-61
Wong, Y. C., & Sanggari, V. (2014). Bioethanol production from sugarcane bagasse using fermentation process. Oriental journal of chemistry, 30(2), 507-513.
Yacob Gebreyohannes Hiben, Y. (2013). Long-term Bioethanol Shift and Transport Fuel Substitution in Ethiopia: Status, Prospects, and Implications.
Zhang, M., Zhu, R., Zhang, M., & Wang, S. (2014). Creation of an ethanol-tolerant Saccharomyces cerevisiae strain by 266 nm laser radiation and repetitive cultivation. Journal of bioscience and bioengineering, 118(5), 508-513.
Zhang, X., Beltranena, E., Christensen, C., & Yu, P. (2012). Use of a dry fractionation process to manipulate the chemical profile and nutrient supply of a coproduct from bioethanol processing. Journal of agricultural and food chemistry, 60(27), 6846-6854.
About this Article
Cite this Article
APA
Admas A., Kiros S., Tafesse M., Tesfaye A., Hawaz E. & Muleta D. Menbere I.P. (2025). Isolating and Characterizing Potential Indigenous Wild Yeasts for Bioethanol Production from Local Resources. SustainE. 1(2) 1-22. https://doi.org/10.55366/suse.v1i2.11
Chicago
Alene Admas, Solomon Kiros, Mesfin Tafesse, Anteneh Tesfaye, Estifanos Hawaz and Diriba Muleta (2025). Isolating and Characterizing Potential Indigenous Wild Yeasts for Bioethanol Production from Local Resources.” SustainE. 1, no. 2 (May 3, 2025). 1-22. https://doi.org/10.55366/suse.v1i2.11
Received
16 January 2025
Accepted
24 March 2025
Published
3 May 2025
Corresponding Author Email: isabellanyambayo@gmail.com
Disclaimer: The opinions and statements expressed in this article are the authors’ sole responsibility and do not necessarily reflect the viewpoints of their affiliated organizations, the publisher, the hosted journal, the editors, or the reviewers. Furthermore, any product evaluated in this article or claims made by its manufacturer are not guaranteed or endorsed by the publisher.
Distributed under Creative Commons CC-BY 4.0
Share this article
Use the buttons below to share the article on desired platforms.














