Oladejo, B. O 1 Bayode, M. T 2 Oladejo, T. C 3 Adejoh, C. A 4 Oluwaluyi, A. O 5 & Olurotimi, B. A 6
1Department of Microbiology, Federal University of Technology, Akure, PMB 704, Ondo State, Nigeria
2Department of Pharmaceutical Microbiology and Biotechnology, University of Medical Sciences, Ondo. P.M.B. 536, Ondo State, Nigeria
*Corresponding Author Email: booladejo@futa.edu.ng
…
Highlights
- Contaminated pumpkin, water leaf, and amaranth green vegetables are vehicles of antibiotic-resistant infectious agents.
- The selected vended vegetables in Ondo City harbour metallo-beta-lactamase, carbapenemase, imipenemase, and oxacillinase resistance genes.
- The Vibrio strains from the selected vended vegetables were multidrug-resistant isolates.
- Beta-lactamase genes could pose a serious threat to beta-lactamase inhibitors.
- Detection of genes such as TEM, SHV, NDM, and VIM highlights the complexity of antibiotic resistance mechanisms.
Abstract
Extended spectrum beta-lactamses (ESBLs) are bacterial enzymes that hydrolyse extended-spectrum cephalosporins, primarily in Gram-negative bacteria. Mutated from TEM-1, TEM-2, or SHV-1 genes, they break down multiple beta-lactam antibiotics through plasmid-encoded mechanisms. This study investigated the prevalence of ESBL genes in Vibrio strains isolated from purposively selected pumpkin, water leaf, and amaranth green vegetables sold in Ondo City, Nigeria, to investigate ESBL genes in Vibrio strains. One hundred vegetable samples were collected and analysed via microbiological and polymerase chain reaction (PCR) to detect specific ESBL genes. The antibiotic sensitivity profile was estimated via disc diffusion following the Clinical Laboratory Standard Institute guidelines. Vibrio cholerae exhibited intermediate resistance to gentamycin (15.17 ± 0.44 mm) and erythromycin (17.00 ± 0.57 mm) but complete resistance to ciprofloxacin (0.00 ± 0.00 mm). V. parahaemolyticus was susceptible to pefloxacin (21.17 ± 0.44 mm) but resistant to ceftriaxone. The TEM and SHV genes were detected in V. cholerae and V. alginolyticus, with band sizes of approximately 258 bp and 318 bp, respectively. However, V. parahaemolyticus did not harbour any detectable ESBL genes. Vibrio species exhibited complex antibiotic resistance profiles, requiring precise antimicrobial selection based on strain-specific susceptibility patterns, which calls for improved food safety measures in urban markets to mitigate the risk of antibiotic-resistant infections transmitted through contaminated vegetables. This study contributes valuable insights into the clinical implications of antibiotic resistance in foodborne pathogens. It highlights the importance of monitoring agricultural practices that may facilitate the spread of beta-lactamase resistance genes.
Keywords: ESBL (extended-spectrum beta-lactamase), Vibrio, Antibiotic resistance, Vegetables, Polymerase chain reaction (PCR).
1. Introduction
The global rise of antibiotic-resistant bacteria has emerged as a significant public health issue, particularly concerning Vibrio species, commonly found in aquatic environments and critical pathogens responsible for foodborne illnesses. A significant cholera outbreak in Lagos State in 2022 demonstrated the potential for transmission of multidrug-resistant V. cholerae through contaminated vegetables, resulting in 2,789 cases and 84 fatalities (Adebayo et al., 2023). Extended spectrum beta-lactamses (ESBLs) are bacterial enzymes that hydrolyse extended-spectrum cephalosporins, primarily in Gram-negative bacteria. Mutated from TEM-1, TEM-2, or SHV-1 genes, they break down multiple beta-lactam antibiotics through plasmid-encoded mechanisms (Bayode et al., 2021). The presence of extended-spectrum beta-lactamases (ESBLs) in these strains poses a serious risk, as these enzymes confer resistance to a broad spectrum of beta-lactam antibiotics, complicating treatment options (Loo & Jonge, 2020; Yamin, 2023). In Ondo City, an urban setting, inadequate hygiene and food safety practices surrounding the vending of local vegetables in public markets can facilitate the transfer of resistant foodborne bacterial strains from the environment or agricultural produce to humans (Olatunji et al., 2023). Olatunji et al. (2023) also observed that market operators in Ondo City face significant infrastructure challenges, with over 76% travelling more than 100 meters for water supply and 85% travelling over 100 meters to access toilet facilities, indicating substantial sanitation and hygiene constraints. Thus, comprehensive studies are urgently needed to assess the prevalence of ESBL-producing Vibrio strains in local vegetable sources.
Okiki et al. (2019) and Umeaku et al. (2022) reported that vended local vegetables, which are often irrigated with untreated water, can serve as reservoirs for pathogenic bacteria, including Vibrio. The identification of ESBL-producing Vibrio strains in these contexts raises concerns regarding potential transmission to consumers, particularly in urban areas where food safety practices may be compromised, as demonstrated by Harris et al. (2023) in their study on the prevalence of antibiotic-resistant bacteria in urban food markets. Various molecular techniques, particularly polymerase chain reaction (PCR), have been utilised to detect ESBL genes in environmental samples; however, data concerning their prevalence in Vibrio strains associated with vegetables are scarce (Naas et al., 2019; Malagón et al., 2020). While molecular techniques like PCR have extensively investigated ESBL genes in vegetable-associated bacterial strains, the critical research gap persists in comprehensively characterising ESBL prevalence, specifically within Vibrio strains, highlighting a significant limitation in the current microbiological understanding of vegetable-associated bacterial resistance mechanisms. Previous investigations by Abodaia et al. (2020) and Ansari et al. (2022) have suggested a link between agricultural practices and the spread of antibiotic resistance, emphasising the necessity for thorough research in urban areas such as Ondo City. The increasing incidence of foodborne illnesses linked to antibiotic-resistant bacteria highlights the urgent need for research into these pathogens’ sources and transmission pathways, as stated by Yamin (2023), who reported significant correlations between food handling practices and the prevalence of resistant strains.
Understanding the prevalence of ESBL genes in Vibrio strains associated with vended local vegetables is essential for effective public health monitoring and intervention strategies. Hence, policy implications for ESBL gene prevalence in Vibrio strains demand comprehensive food safety interventions, including implementing rigorous agricultural water quality standards, enhancing market hygiene protocols, establishing mandatory bacterial screening for vegetable vendors, and developing targeted surveillance systems to track antimicrobial resistance transmission in food supply chains. Despite increasing concerns regarding antibiotic resistance in foodborne pathogens, comprehensive data on ESBL genes in Vibrio strains linked to local vegetables remain limited. This knowledge gap poses a significant public health risk, especially in locations such as Ondo City, where vegetables constitute a dietary staple. Without a clear understanding of the public health threat posed to food consumers, effective strategies to combat the proliferation of antibiotic-resistant bacteria cannot be developed.
The aim of this study is to address a significant gap in the literature by examining how local agricultural practices contribute to the spread of resistant Vibrio strains in local vegetables sold in public markets in Ondo City. The findings could offer critical insights for policymakers and public health officials in formulating targeted interventions to mitigate the risks associated with food consumption to detect and characterise extended-spectrum beta-lactamase genes in Vibrio strains associated with publicly sold local vegetables.
2. Materials and Methods
2.1 Sample collection
One hundred vegetable samples (pumpkin, water leaf, and amaranth green) were purchased from different markets in Ondo City, Nigeria, via sterile collection bags. The samples were transported to the Microbiology Laboratory at the University of Medical Sciences (UNIMED) teaching hospital Odochida, Ondo, within 1 hour for microbiological analysis. Pumpkin, water leaf, and amaranth green were selected due to their significant local dietary importance. These vegetables are commonly consumed in Ondo City, are widely available in local markets, and represent staple vegetable varieties in the Nigerian diet. Approximately 33 samples of pumpkin, 33 samples of water leaf and 34 samples of amaranth green vegetables were purposively collected from the Oja-oba market, the Adeyemi University College market and the Odochida market in Ondo City. The selection of markets is justified by the need to assess food safety and contamination levels in cities where traditional market practices and settings often lead to high levels of foodborne bacterial pathogens.
2.2 Isolation of Vibrio species
Being the primary plating medium universally utilised for Vibrio culture, thiosulfate citrate bile salt sucrose (TCBS) agar was used for Vibrio isolation. The edible portions of the vegetable samples were separated, and 150–200 g of each sample was rinsed with 200 mL of distilled water. Serial dilutions (10-4 and 10-5) were prepared, and 1 mL of each dilution was inoculated onto TCBS agar plates via the pour plate method. The samples were subsequently incubated at 37°C for 48 hours. Vibrio colonies (yellow or green) were further subcultured on TCBS agar for confirmation, which enabled precise detection of Vibrio species through selective medium characteristics, differential colour changes, and adherence to standard microbiological characterisation protocols, ultimately minimising contamination risks and ensuring reliable bacterial enumeration. Colonies in the media were enumerated and expressed as CFU/g. Microbial isolates were characterised via culture, microscopic, and biochemical methods via standard manuals.
2.3 Molecular identification of Vibrio strains
DNA was extracted from the Vibrio isolates via cell lysis, protein precipitation, and DNA precipitation. PCR amplification of the 16S rRNA gene was performed via GoTaq® Green Master Mix (Promega, USA) and the universal primers 27F and 1525R in a GeneAmp 9700 PCR System (Applied Biosystems, USA). The amplified products were visualised on 1% agarose gels, purified via ethanol precipitation, and sequenced via an Applied Biosystems 3130xl Genetic Analyser (USA).
This study used the primers 27F and 1525R as universal bacterial primers with specific characteristics for 16S rRNA gene amplification. 27F (Forward) targets the conserved region at the 5′ end of the bacterial 16S rRNA gene. 1525R (Reverse) targets the complementary region at the 3′ end of the bacterial 16S rRNA gene. These are extensively validated across multiple bacterial genera, including Vibrio species and recommended in numerous microbiological studies for broad-spectrum bacterial identification. The negative PCR control used was nuclease-free water (Thermo Fisher Scientific, MA, USA), while the positive PCR control with a known bacterial DNA template used (QIAGEN, USA) ensured reliability and eliminates potential contamination risks. The universal nature of these primers enables comprehensive bacterial identification across diverse species, making them a robust choice for molecular characterisation of Vibrio isolates.
2.4 Inoculum calibration and antibiotic susceptibility test of Vibrio isolates
A loop of test bacterial isolates was inoculated on nutrient broth and incubated for 24 h. Approximately 0.2 mL from the 24-h broth culture of the bacteria was dispensed into 20 mL of sterile nutrient broth and incubated for 3 to 5 h to standardise the culture to 0.5 McFarland standards (106 CFU/mL) before use. Vibrio isolates were tested for antibiotic susceptibility via the Kirby–Bauer disk diffusion method on Mueller‒Hinton agar, following CLSI guidelines (CLSI, 2019). Commercially available antibiotic discs (6 mm diameter) containing 12 different antimicrobial agents were used. Pefloxacin (10 µg), gentamycin (10 µg), ampiclox (30 µg), amoxicillin (30 µg), ceftriaxone (30 µg), ciprofloxacin (10 µg), streptomycin (30 µg), erythromycin (30 µg), cotrimoxazole (30 µg), chloramphenicol (30 µg), sparfloxacin (10 µg), and augmetin (30 µg) were added to the culture plates and incubated at 35 ± 2°C for 24 h. Zones of inhibition were measured and interpreted via CLSI standards. The antibiotic discs were likely sourced from commercial manufacturers like TM Media Antibiotic Discs (New Delhi, India), which produce standardised 6 mm diameter discs aligned with CLSI guidelines. The antibiotic selection rationale includes fluoroquinolones (pefloxacin, ciprofloxacin), covers aminoglycosides (gentamycin, streptomycin), includes beta-lactam antibiotics (amoxicillin, ceftriaxone), all representing commonly used clinical antibiotics in regional treatment protocols. The selected antibiotics were strategically chosen to provide a comprehensive antimicrobial susceptibility profile for Vibrio species, enabling detailed resistance pattern analysis consistent with standard microbiological testing protocols
2.5 Detection of beta-lactamase genes in Vibrio species
Polymerase chain reaction (PCR) was performed to detect beta-lactamase resistance genes in the selected Vibrio isolates (Table 1). The reaction mixture contained 5X PCR SYBR Green buffer (Thermo Fisher Scientific, USA), MgCl2, dNTPs, forward and reverse primers, Taq DNA polymerase, and template DNA. The amplified products were visualised on 1% agarose gels. Molecular investigations of beta-lactamase antibiotic resistance genes in selected isolates of V. cholerae, V. parahaemolyticus, and V. alginolyticus were conducted via simple PCR on extracted DNA with gene-specific primers (Di Maro et al., 2024).
The selected beta-lactamase genes represent critical molecular mechanisms of antibiotic resistance in Vibrio species, with global and regional significance, such as Metallo-beta-lactamases (VIM and NDM), conferring broad resistance to carbapenem antibiotics. Another metallo-beta-lactamase (IMP) with widespread resistance potential. Carbapenemase gene (KPC) associated with high-level antibiotic resistance. Oxacillinase gene (OXA) enabling resistance to extended-spectrum beta-lactams. Temoneira (TEM) and Sulfhydryl Variable (SHV) are classic extended-spectrum beta-lactamase genes amplified via PCR in this study. The varied annealing temperatures (47-50°C) reflect specific primer-template interactions. The lower temperatures (47-49°C) enable more flexible primer binding, optimised to balance specificity and amplification efficiency and are consistent with standard PCR protocols for beta-lactamase gene detection.
Table 1: DNA primer sets used for the detection of beta-lactamase genes in Vibrio strains in vegetables
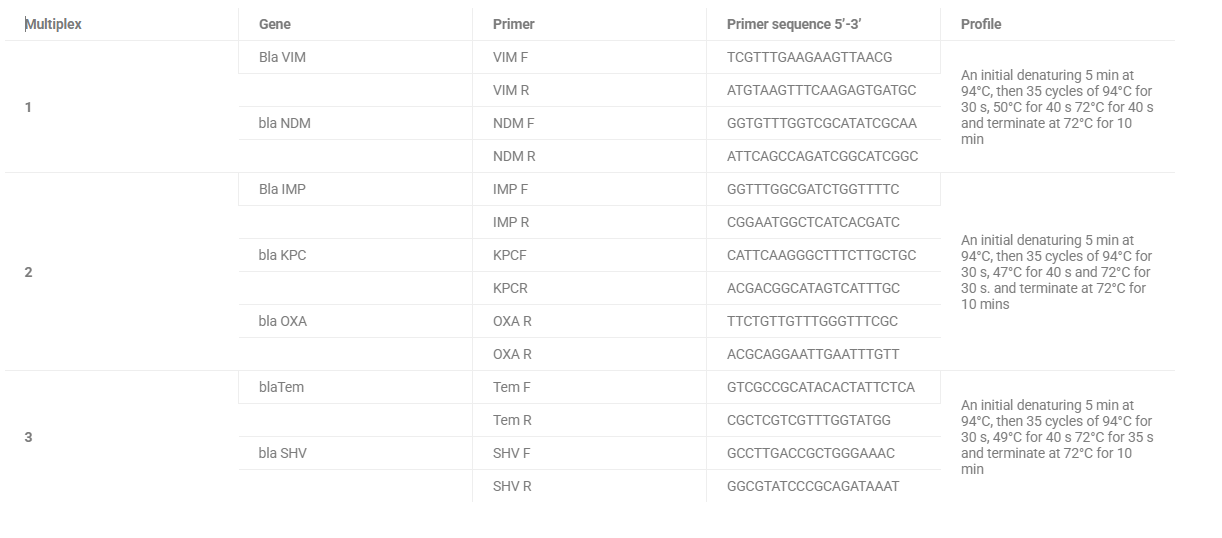
| Multiplex | Gene | Primer | Primer sequence 5’-3’ | Profile |
| 1 | Bla VIM | VIM F | TCGTTTGAAGAAGTTAACG | An initial denaturing 5 min at 94°C, then 35 cycles of 94°C for 30 s, 50°C for 40 s 72°C for 40 s and terminate at 72°C for 10 min |
| VIM R | ATGTAAGTTTCAAGAGTGATGC | |||
| bla NDM | NDM F | GGTGTTTGGTCGCATATCGCAA | ||
| NDM R | ATTCAGCCAGATCGGCATCGGC | |||
| 2 | Bla IMP | IMP F | GGTTTGGCGATCTGGTTTTC | An initial denaturing 5 min at 94°C, then 35 cycles of 94°C for 30 s, 47°C for 40 s and 72°C for 30 s. and terminate at 72°C for 10 mins |
| IMP R | CGGAATGGCTCATCACGATC | |||
| bla KPC | KPCF | CATTCAAGGGCTTTCTTGCTGC | ||
| KPCR | ACGACGGCATAGTCATTTGC | |||
| bla OXA | OXA R | TTCTGTTGTTTGGGTTTCGC | ||
| OXA R | ACGCAGGAATTGAATTTGTT | |||
| 3 | blaTem | Tem F | GTCGCCGCATACACTATTCTCA | An initial denaturing 5 min at 94°C, then 35 cycles of 94°C for 30 s, 49°C for 40 s 72°C for 35 s and terminate at 72°C for 10 min |
| Tem R | CGCTCGTCGTTTGGTATGG | |||
| bla SHV | SHV F | GCCTTGACCGCTGGGAAAC | ||
| SHV R | GGCGTATCCCGCAGATAAAT |
Gel electrophoresis of beta-lactamase genes from Vibrio strains isolated from selected vended vegetables
A 1% agarose gel was prepared via 1X TAE buffer (Thermo Fisher Scientific, Waltham, MA, USA) in a Panasonic microwave oven (Kadoma, Osaka, Japan) to confirm the integrity of the amplified gene fragments. The molten agarose was allowed to cool to 60°C and stained with 3 μL of 0.5 μg/mL ethidium bromide solution (Thermo Fisher Scientific, Waltham, MA, USA). A comb was inserted into the slots of a casting tray (Bio-Rad, Hercules, CA, USA), and the stained agarose was poured into the tray. After solidifying for 20 minutes, the gel was placed in a gel tank (Bio-Rad) and submerged in 1X TAE buffer. Two microliters of 10X blue gel loading dye (Thermo Fisher Scientific, Waltham, MA, USA) was added to 4 μL of each PCR product and loaded into the wells after a 100 bp DNA ladder (Thermo Fisher Scientific) was loaded into the first well, combined with 2 μL of 10X blue gel loading dye, totalling 6 μL per sample. The gel was electrophoresed at 120 V for 45 minutes using a PowerPac HC power supply (Bio-Rad). The DNA bands were visualised under UV light using a ChemiDoc MP imaging system (Bio-Rad) and photographed. The sizes of the PCR products were estimated by comparing their mobility with that of the 100 bp molecular weight (marker) ladder (Miao et al., 2022).
3. Results and Discussion
Morphological characterisation of presumptive Vibrio sp. on TCBS agar
Green and yellow colonies were observed on thiosulfate‒bile salt‒sucrose (TCBS) agar plates. On TCBS, yellow colonies were presumptively identified as V. alginolyticus or V. cholerae, whereas green or blue-green colonies were assumed to be V. parahaemolyticus, as shown in Table 2.
Table 2: Morphological characterisation of Vibrio colonies on TCBS
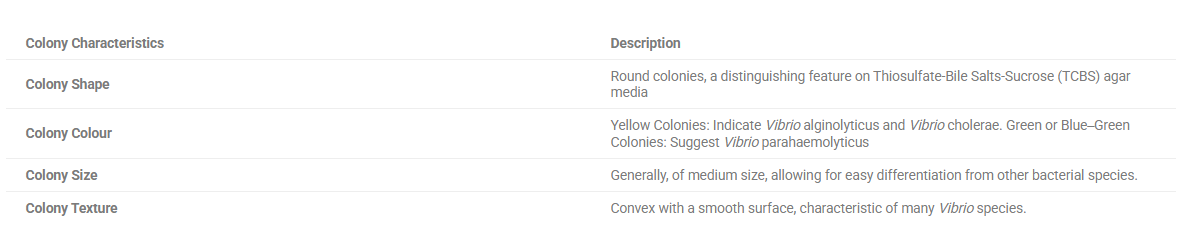
| Colony Characteristics | Description |
| Colony Shape | Round colonies, a distinguishing feature on Thiosulfate-Bile Salts-Sucrose (TCBS) agar media |
| Colony Colour | Yellow Colonies: Indicate Vibrio alginolyticus and Vibrio cholerae. Green or Blue‒Green Colonies: Suggest Vibrio parahaemolyticus |
| Colony Size | Generally, of medium size, allowing for easy differentiation from other bacterial species. |
| Colony Texture | Convex with a smooth surface, characteristic of many Vibrio species. |
3.1 Cultural identity of Vibrio species from selected vended vegetables
Gram-negative short-rod bacteria were isolated from vegetables sold at public markets in Ondo City and were presumptively identified as V. alginolyticus, V. cholerae or V. parahaemolyticus, as illustrated in Table 3. Vibrio parahaemolyticus showed a high prevalence of 37 %, followed by V. cholerae (35 %) and V. alginolyticus (28 %) as least prevalent.
Table 3: Cultural characteristics of Vibrio species isolated from selected vended vegetables
| Sample ID | Isolates | Max Score | Total Score | Query Cover | % Identity | Accession No. |
| 1 | V. cholerae | 2567 | 2567 | 99% | 99.93 | OP019729.1 |
| 2 | V. parahaemolyticus | 2630 | 2630 | 100% | 100.00 | NR_041838.1 |
| 3 | V. alginolyticus | 2518 | 2518 | 100% | 100.00 | FN436276.1 |
Key: + = positive, – = negative, -ve = negative
3.2 Molecular identity of Vibrio strains from selected vended vegetables
The gel image shows the purified DNA bands of three Vibrio species: V. cholerae (lane 1), V. parahaemolyticus (lane 2), and V. alginolyticus (lane 3). The molecular weight ladder of 1500 base pairs (bp) (MK) was also included for size reference (Figure 1). The 1500 bp molecular weight ladder was strategically selected to provide comprehensive size reference for the Vibrio species gene fragments because it covers the molecular weight range of most bacterial gene amplicons, enables precise size estimation for typical PCR products, and provides multiple reference points between 100-1500 bp. It is also sufficiently broad range to capture most 16S rRNA and beta-lactamase gene fragments, allows accurate molecular weight determination for Vibrio species genetic material and consistent with recommended DNA marker standards for bacterial molecular studies. The NCBI BLAST results in Table 4 show the sequence identity of the study’s Vibrio isolates isolated from vegetables.

Figure 1: Electrophoretic gel image showing the purified DNA bands of Vibrio strains
Key: MK- Molecular weight ladder; 1- V. cholerae; 2- V. parahaemolyticus; 3- V. alginolyticus
Table 4: NCBI BLAST showing the sequence identity of the Vibrio isolates
| Sample ID | Isolates | Max Score | Total Score | Query Cover | % Identity | Accession No. |
| 1 | V. cholerae | 2567 | 2567 | 99% | 99.93 | OP019729.1 |
| 2 | V. parahaemolyticus | 2630 | 2630 | 100% | 100.00 | NR_041838.1 |
| 3 | V. alginolyticus | 2518 | 2518 | 100% | 100.00 | FN436276.1 |
3.3 Antibiogram of Vibrio strains isolated from selected vended vegetables
Vibrio cholerae presented intermediate resistance to gentamycin (15.17±0.44 mm), erythromycin (17.00 ± 0.57 mm), spasfloxacin (15.00±0.58 mm), and amoxicillin/clavulanic acid (15.50±0.29 mm) but resistance to critical antibiotics, including ciprofloxacin (0.00±0.0 mm), amoxicillin (0.00±0.0 mm), and chloramphenicol (13.00± 0.57 mm), which are often used to treat cholera. V. parahaemolyticus was susceptible to perfloxacin (21.17±0.44 mm) and spasfloxacin (20.00±0.58 mm), indicating potential treatment options, while it was resistant to several antibiotics, most notably ciprofloxacin (7.00±0.5 mm) and ceftriaxone (0.00±0.0 mm). V. alginolyticus was susceptible to Ampicillin (20.00±0.57 mm) and chloramphenicol (19.00±1.15 mm), which could be effective treatment options, but was resistant to multiple antibiotics, including complete resistance to perfloxacin (0.00±0.0 mm) and ciprofloxacin (0.00±0.0 mm) (Table 5). Intermediate antibiotic resistance in Vibrio species represents a critical clinical challenge, signalling reduced drug effectiveness, potential treatment complications, and increased risk of bacterial adaptation, which necessitates careful antibiotic selection, dosage adjustment, and sophisticated diagnostic strategies to mitigate the evolving antimicrobial resistance mechanisms.
Table 5: Antibiogram of Vibrio strains isolated from selected vended vegetables

The values are expressed as the means ± S.E.ss (n=3). The means were separated via DNMRT (P>0.05). The values along the same row that share the same superscript are not significantly different.
Keys: Iso. – Isolates; V. c. – Vibrio cholerae; V. p. – Vibrio parahaemolyticus; V. a. – Vibrio alginolyticus; PEF- Perfloxacin (30 mg); GEN- Gentamycin (10 mg); AMP-Ampiclox (30mg); AMX- Amoxicillin (30 mg); CEF – Ceftriaxone (30 mg); CIP- Ciprofloxacin (10 mg); STP- Streptomycin (30 mg); ERY- Erythromycin (10 mg); COT- Trimethoprim-Sulfamethoxazole (30 mg); Chloramphenicol (30 mg); SPA – Sparfloxacin (30 mg); AUG – Amoxicillin/Clavulanic acid (30 mg)
3.4 Detection of ESBL TEM (Temoneira) and SHV (Sulfhydryl Variable) genes in Vibrio strains from selected vended vegetables
Agarose gel electrophoresis of the PCR products of the ESBL TEM and SHV genes amplified from selected Vibrio isolates revealed DNA band sizes of approximately 258 base pairs (bp) and 318 bp, indicating the presence of TEM and SHV, respectively. The gel image indicates a positive DNA band of TEM for V. cholerae and V. alginolyticus, but none of the ESBL genes were detected for V. parahaemolyticus (Figure 2).

Figure 2: Amplified PCR products of ESBL-producing TEM and SHV genes from selected vended vegetables
Keys: MK- Molecular weight ladder; 1- V. cholerae; 2- V. parahaemolyticus; 3- V. alginolyticus
3.5 Detection of the ESBL NDM (New Delhi metallo-beta-lactamase) and VIM (Verona Integron-encoded metallo-beta-lactamase) genes in Vibrio strains from selected vended vegetables
Agarose gel electrophoresis of the PCR products of the ESBL NDM and VIM genes amplified from selected Vibrio isolates revealed DNA band sizes of approximately 624 base pairs (bp) and 502 bp, indicating the presence of NDM and VIM, respectively. The gel image indicates positive DNA bands of NDM for V. cholerae and V. parahaemolyticus, and VIM was detected only in the V. cholerae strain (Figure 3).

Figure 3: Amplified PCR products of ESBL-producing NDM and VIM genes from selected vended vegetables
Keys: MK- Molecular weight ladder; 1- V. cholerae; 2- V. parahaemolyticus; 3- V. alginolyticus
4.6 Detection of ESBL KPC (Klebsiella pneumoniae carbapenemase), IMP (imipenemase) and OXA (oxacillinase) genes in Vibrio strains from selected vended vegetables
Agarose gel electrophoresis of the PCR products of the ESBL KP, IMP and OXA genes amplified from selected Vibrio isolates revealed DNA band sizes of approximately 498 base pairs (bp), 568 bp and 190 bp, indicating the KPC IMP and OXA genes, respectively. The gel image indicates a positive DNA band of OXA in all the Vibrio strains, while KPC and IMP were not detected at all (Figure 4).

Figure 4: Amplified PCR products of ESBL-producing KPC IMP and OXA genes from selected vended vegetables
Keys: MK- Molecular weight ladder; 1- V. cholerae; 2- V. parahaemolyticus; 3- V. alginolyticus
3.7 Detection of the ESBL CTM-M (Cefotaxime Munich) gene in Vibrio strains from selected vended vegetables
Agarose gel electrophoresis of the PCR products of the ESBL CTX-M gene amplified from selected Vibrio isolates revealed a band size of approximately 598 bp. The gel image indicates the presence of the CTX-M gene in only V. alginolyticus (Figure 5).

Figure 5: Amplified PCR products of the ESBL-producing CTX-M gene in selected vended vegetables
Keys: MK- Molecular weight ladder; 1- V. cholerae; 2- V. parahaemolyticus; 3- V. alginolyticus
4. Discussion
This study was conducted to detect extended-spectrum beta-lactamase genes in Vibrio strains associated with publicly sold local vegetables. The cultural identities of the Vibrio isolates from this study are analogous to the results of Igbinosa et al. (2021), who carried out biochemical identification of some gram-negative rod Vibrio strains from fresh vegetable samples.
Vibrio parahaemolyticus showed a high prevalence of 37 %, consistent with the tabular data (Table 3). A similar observation of microbial counts in the study by Igbinosa & Odjadjare (2016) in their study in Benin City, Edo State, Nigeria, on the characterisation of antibiogram susceptibility profile of Vibrio species isolated from fresh vegetables ranged from 1.5 × 105 to 3.7 × 105 CFU/g across different vegetable types. These Vibrio species detected from the vegetables in this study represented potential public health risks due to their pathogenic characteristics, as evident in the findings of Igbinosa & Odjadjare (2016) and Igbinosa et al. (2021). The reported prevalence percentages align with broader research indicating that multiple Vibrio species coexist in vegetable ecosystems, with slight variations in their relative abundance. The close distribution (28-37%) suggests a complex microbial landscape in urban market vegetables, emphasising the need for rigorous food safety monitoring, as reported by Igbinosa & Odjadjare (2016) and Igbinosa et al. (2021). This highlighted how Vibrio species could harbour antibiotic resistance genes and virulence determinants, further underscoring the significance of their presence in food supply chains.
The presence of distinct, well-resolved DNA bands for each Vibrio species indicates that the DNA extraction and purification procedures were successful. The band sizes and patterns suggest that these Vibrio strains possess genomic differences that can be differentiated through this gel electrophoresis approach. The gel image provides initial confirmation of the identity of the Vibrio isolates obtained from the vegetable samples collected in Ondo Town. The clear separation of the DNA bands for the three Vibrio species demonstrates the utility of this molecular technique in distinguishing between closely related bacteria.
All three Vibrio species presented multidrug resistance patterns, with V. cholerae being the most resistant, showing no full susceptibility to any tested antibiotic, whereas V. parahaemolyticus and V. alginolyticus each presented susceptibility to two antibiotics, highlighting the need for careful antibiotic selection based on species identification and susceptibility testing. The observed resistance to ciprofloxacin across all three Vibrio species (V. cholerae: 0.00±0.00 mm, V. parahaemolyticus: 7.00±0.5 mm, V. alginolyticus: 0.00±0.00 mm) is particularly concerning, given its common use in treating Vibrio infections, suggesting the need for alternative treatment strategies in Ondo city.
The detection of extended-spectrum beta-lactamase (ESBL) genes, specifically TEM and SHV, in Vibrio cholerae and V. alginolyticus, but not in V. parahaemolyticus, highlights the varying resistance profiles among these species. The presence of the NDM and VIM genes in Vibrio cholerae and V. parahaemolyticus suggests that these strains have the ability to resist treatment with beta-lactam antibiotics, which are commonly used to treat bacterial infections. This finding is consistent with that of De et al. (2021), indicating that V. cholerae has emerged as a multidrug-resistant pathogen that often harbours multiple resistance genes, including those encoding ESBLs. The absence of these genes in V. parahaemolyticus could indicate that different evolutionary pressures or environmental factors influence resistance development, as observed by Abdelaziz-Gobarah et al. (2022), who reported that environmental Vibrio species may exhibit distinct resistance mechanisms on the basis of their habitats. The detection of the OXA gene across all three species further highlights the adaptability of these bacteria in the acquisition of resistance mechanisms, which can complicate treatment options for infections caused by these pathogens. The evolving landscape of Vibrio species antibiotic resistance reveals a complex global trend characterised by widespread multidrug resistance, with the study by Pandey et al. (2023) demonstrating that 80% of isolates exhibit resistance to beta-lactam antibiotics and their derivatives and multiple antibiotic resistance (MAR) indices consistently exceeding the recommended 0.2 threshold, highlighting the urgent need for sophisticated molecular surveillance and adaptive treatment strategies to mitigate the potential public health risks associated with these rapidly adapting bacterial populations.
Moreover, PCR amplification of the ESBL TEM and SHV genes in the selected Vibrio isolates indicated the presence of specific DNA band sizes corresponding to the TEM and SHV genes (Amalina et al., 2019). This differential distribution of ESBL genes among Vibrio species highlights the genetic diversity and variability in antibiotic resistance determinants within these bacterial populations. Wang et al. (2020) reported varying patterns of ESBL gene carriage in Vibrio species, emphasising the complex nature of antibiotic resistance gene dissemination. This distinct distribution of carbapenemase genes (NDM and VIM) among the Vibrio species suggests unique genetic backgrounds and potential differences in antibiotic resistance profiles.
Previous research has highlighted the emergence of carbapenem resistance in Vibrio species, which poses challenges for clinical treatment and public health management, as affirmed by El-Zamkan et al. (2023). The differential distribution of beta-lactamase genes (KPC, IMP, and OXA) underscores the diverse genetic repertoire of antibiotic resistance determinants in Vibrio species, emphasising the importance of surveillance and monitoring of resistance mechanisms in environmental isolates. Di Maro et al. (2024) reported the role of beta-lactamases in conferring resistance to multiple antibiotics, which poses a significant threat to antimicrobial therapy. The presence of these ESBL genes is alarming, as they confer resistance to important beta-lactam antibiotics. The detection of NDM (New Delhi metallo-beta-lactamase) is particularly concerning because of its association with multidrug resistance. These findings align with the study of Adebayo et al. (2023), who observed a significant cholera outbreak in Lagos State in 2022 which underlines the potential for transmission of multidrug-resistant V. cholerae through contaminated vegetables, hence strengthening the potential emergence of ESBL-producing Vibrio species in Ondo city, and globally as reviewed by Kakoullis et al. (2023) observing the increasing prevalence of ESBL-producing V. cholerae.
The molecular investigation via PCR of Vibrio species in Ondo city reveals a complex antibiotic resistance landscape characterised by widespread OXA gene prevalence across all strains, selective CTX-M gene presence in V. alginolyticus detected in the vegetables investigated in this study, and implications for treatment strategies that necessitate species-specific approaches, reflecting regional genetic adaptation mechanisms and environmental selective pressures that underscore the critical need for sophisticated diagnostic interventions to address the evolving multidrug resistance patterns in bacterial populations.
The molecular identity findings revealed a complex antibiotic resistance profile across Vibrio species, with V. cholerae demonstrating the most diverse resistance gene repertoire (TEM, NDM, VIM, and OXA genes), V. alginolyticus exhibiting limited but significant resistance (TEM, CTX-M, and OXA genes), and V. parahaemolyticus displaying selective resistance mechanisms (NDM and OXA genes), which directly correlate with varied antibiotic susceptibility patterns, including V. cholerae intermediate and complete resistance to multiple antibiotics, V. parahaemolyticus selective susceptibility, and V. alginolyticus resistance to critical antibiotics, ultimately underscoring the intricate relationship between genetic resistance determinants and phenotypic antibiotic resistance mechanisms that necessitate sophisticated diagnostic and treatment strategies. Recent studies by Adesiyan et al. (2022) and Carole and Abubakar (2024) on Vibrio species reveal a complex landscape of antibiotic resistance mechanisms, with molecular genetic findings directly correlating to phenotypic resistance patterns. Findings by Carole and Abubakar (2024) also indicate that 97% of Vibrio isolates exhibit multiple antibiotic resistance indices above the 0.2 thresholds, with prevalent resistance patterns observed against erythromycin (95%), sulphamethoxazole (94%), and rifampicin (92%). The molecular identity findings demonstrate diverse resistance gene profiles across different Vibrio species, with V. cholerae showing the most comprehensive resistance gene repertoire, including TEM, NDM, VIM, and OXA genes (Adesiyan et al., 2022). Notably, 52.9% of isolates were found to be multidrug-resistant, with multiple antibiotic resistance indices reaching up to 1.0, underscoring the critical need for sophisticated diagnostic and treatment strategies to mitigate the evolving antimicrobial resistance mechanisms in these potentially hazardous bacterial species.
5. Conclusion
The detection of specific resistance genes, such as TEM, SHV, NDM, and VIM, across these species underscores the complexity of antibiotic resistance mechanisms and the potential for multidrug resistance, particularly in Vibrio cholerae and Vibrio alginolyticus. The absence of certain resistance genes in Vibrio parahaemolyticus suggests differing evolutionary pressures, indicating that environmental factors play crucial roles in shaping resistance profiles. These findings raise significant concerns regarding food safety and public health, as the presence of these resistant strains in vegetables could pose risks to consumers as well as serve as a reservoir for antibiotic-resistant bacteria. The comprehensive mitigation strategy for addressing Vibrio species antibiotic resistance demands a multifaceted approach encompassing stringent policy interventions, including mandatory molecular screening of food products, implementing national antibiotic stewardship guidelines, regulating agricultural and aquaculture antibiotic usage, developing advanced surveillance systems tracking resistance gene evolution, and promoting alternative treatment modalities such as targeted vaccination protocols and phytotherapeutic interventions, with the ultimate goal of reducing horizontal gene transfer potential, minimising environmental antibiotic exposure, and enhancing molecular diagnostic capabilities to effectively combat the complex ecological dynamics of bacterial adaptation and resistance mechanisms that pose significant public health risks.
6. Acknowledgement
The authors are grateful to the Department of Microbiology, University of Medical Sciences, Ondo, Ondo State, Nigeria, for providing the technical support needed for the study.
Conflict of Interest
The authors declare no conflict of interest.
References
Abdelaziz Gobarah DE, Helmy SM, Mahfouz NB, Fahmy HA, Abou Zeid MAEHM (2022). Virulence genes and antibiotic resistance profile of Vibrio species isolated from fish in Egypt. Veterinary Research Forum. 13(3):315-321.
Abodaia, A., Abo-Nasr, A., & El-Sayed, M. (2020). The impact of agricultural practices on the spread of antibiotic resistance among foodborne pathogens. Environmental Science and Pollution Research, 27(10), 11645-11656.
Adebayo. O., Olanipekun, A. J., Ojo, O. (2023). Outbreak of multidrug-resistant Vibrio cholerae in Lagos State, Nigeria: A public health concern. Journal of Infectious Diseases, 227(4), 123–130.
Adesiyan, I.M., Bisi-Johnson, M.A. & Okoh, A.I. (2022). Incidence of antibiotic resistance genotypes of Vibrio species recovered from selected freshwaters in Southwest Nigeria. Sci Rep 12, 18912. https://doi.org/10.1038/s41598-022-23479-0
Ansari, S., Kumar, V., Bhatt, D. N., Irfan, M., & Datta, A. (2022). N-acetylglucosamine sensing and metabolic engineering for attenuating human and plant pathogens. Bioengineering, 9(2), 64.
Bayode, M., Olalemi, A., Oladejo, B. (2021). Multiple Antibiotic Resistant Index and Detection of QnrS and QnrB Genes in Bacterial Consortium of Urine Samples from Clinical Settings. European Journal of Biological Research, 11, 45-56.
Carole, N.Y.M, and Abubakar, U. (2024). Study on Antibiotic Resistance Profiles Exhibited by Vibrio Species Isolated from Landfill Soils in Zaria Metropolis, Northern Nigeria. Umaru Musa Yardua University, Conference Special Issue. 9 (3): 252 – 258. https://doi.org/10.47430/ujmr.2493.031
CLSI. (2019). Performance Standards for Antimicrobial Susceptibility Testing (29th ed.). Clinical and Laboratory Standards Institute.
Di Maro, O., Proroga, Y. T., Castellano, S., Balestrieri, A., Capuano, F., Arletti, E., Vietina, M., Bizzarri, M., Murru, N., Peruzy, M. F. & Cristiano, D. (2024). Detection of pathogenic Vibrio spp. in foods: Polymerase chain reaction-based screening strategy to rapidly detect pathogenic Vibrio parahaemolyticus, Vibrio cholera and Vibrio vulnificus in Bivalve Molluscs and preliminary results. Italian Journal of Food Safety, 13(1):11635
De, R. (2021). Mobile genetic elements of Vibrio cholerae and the evolution of its antimicrobial resistance. Frontiers in Tropical Diseases. 2: 691604.
El-Zamkan, M. (2023). Molecular characterisation of Vibrio species isolated from dairy and water samples. Scientific Reports. 13(1).
Harris, J., Smith, R., & Jones, L. (2023). Prevalence of antibiotic-resistant bacteria in urban food markets: Implications for public health. Food Control, 143, 109239.
Igbinosa, E. O., & Odjadjare, E. E. (2016). Characterisation of antibiogram susceptibility profile of Vibrio species isolated from fresh vegetables. African Scientist, 2016: 17(2).
Igbinosa, E. O., Beshiru, A., Igbinosa, I. H., Ogofure, A. G., & Uwhuba, K. E. (2021). Prevalence and characterisation of foodborne Vibrio parahaemolyticus from African salad in Southern Nigeria. Frontiers in microbiology, 12: 632266.
Kakoullis, L., Papachristos, A., Karaolia, P., Tzioni, E., Panayiotou, C., Paphitou, N. I., & Papageorgiou, D. K. (2023). Extended-Spectrum β-Lactamases (ESBL): Challenges and Opportunities. Biomedicines. 11(11): 2937.
Loo, M. P. J., & Jonge, E. (2020). Data validation: Ensuring the reliability of data sets for research purposes. In Handbook of Statistical Data Editing and Imputation. (pp. 1-15). Wiley.
Malagón, M., González-López, J. J., & Rodríguez-Baño, J. (2020). Detection of extended-spectrum beta-lactamase genes in environmental samples: A review of molecular techniques. Journal of Antimicrobial Chemotherapy, 75(2), 257-267.
Miao, G., Zhang, L., Zhang, J., Ge, S., Xia, N., Qian, S., & Qiu, X. (2020). Free convective PCR: From principle study to commercial applications—A critical review. Analytica Chimica Acta, 1108: 177-197.
Naas, T., Nordmann, P., & Poirel, L. (2019). Emergence of extended-spectrum beta-lactamases in Vibrio spp.: A global perspective. Clinical Microbiology Reviews, 32(4), e00045-19.
Okiki, P. A., Adesanya, O., & Ojo, O. (2019). Vended local vegetables as reservoirs for pathogenic bacteria: Implications for public health in Nigeria. Journal of Food Safety, 39(2), e12534.
Olatunji, S. A., Yoade, A. O., Atoyebi, O. S. (2023). Environmental Sanitation Practices in Traditional Markets of Sub-Saharan Africa. Journal of Applied Sciences and Environmental Management, 27(12), 2949-2456.
Pandey, R., Sharma, S., Sinha, K. K. (2023). Evidence of Antibiotic Resistance and Virulence Factors in Environmental Isolates of Vibrio Species. Antibiotics (Basel). 12(6), 1062. doi: 10.3390/antibiotics12061062
Umeaku, N., Nwankwo, C., & Okwuosa, C. (2022). Assessment of bacterial contamination in vended vegetables: A case study from Nigeria. African Journal of Microbiology Research, 16(5), 185-193.
Wang, H., Lin, S., Mohapatra, A., Kumar, R., & Wang, H. (2020). A review of the functional annotations of important genes in the ahpnd-causing pva1 plasmid. Microorganisms. 8(7): 996.
Yamin, M. (2023). Antibiotic resistance among foodborne pathogens: A growing concern for public health safety. Foodborne Pathogens and Disease, 20(1), 1–10.
About this Article
Cite this Article
APA
Oladejo, B. O., Bayode, M. T., Oladejo, T. C., Adejoh, C. A., Oluwaluyi, A. O. & Olurotimi, B. A.(2025). Detection of Extended Spectrum Beta-Lactamase Genes in Vibrio Strains Associated with Selected Local Vegetables Sold in Public Markets in Ondo City, Nigeria. In Akinyele B.J., Kayode R. & Akinsemolu A.A. (Eds.), Microbes, Mentorship, and Beyond: A Festschrift in Honour of Professor F.A. Akinyosoye. SustainE
Chicago
Oladejo, B. O., Bayode, M. T., Oladejo, T. C., Adejoh, C. A., Oluwaluyi, A. O. and Olurotimi, B. A. 2025. “Detection of Extended Spectrum Beta-Lactamase Genes in Vibrio Strains Associated with Selected Local Vegetables Sold in Public Markets in Ondo City, Nigeria” In Microbes, Mentorship, and Beyond: A Festschrift in Honour of Professor F.A. Akinyosoye, edited by Akinyele B.J., Kayode R. and Akinsemolu A.A., SustainE.
Received
20 November 2024
Accepted
10 January 2025
Published
4 February 2025
Corresponding Author Email: booladejo@futa.edu.ng
Disclaimer: The opinions and statements expressed in this article are the authors’ sole responsibility and do not necessarily reflect the viewpoints of their affiliated organizations, the publisher, the hosted journal, the editors, or the reviewers. Furthermore, any product evaluated in this article or claims made by its manufacturer are not guaranteed or endorsed by the publisher.
Distributed under Creative Commons CC-BY 4.0
Share this article
Use the buttons below to share the article on desired platforms.














