Oluwole, O. R;1Olalemi, A. O.;2 & Akinyele, B. J3
1Department of Basic Science, Federal College of Agriculture, Akure, P.M.B 724, Akure, Ondo State, Nigeria.
2Department of Microbiology, Faculty of Science, Federal University Oye-Ekiti, Ekiti State, Nigeria.
*Corresponding Author Email: omowunmi.oluwole@gmail.com …
Abstract
Aflatoxins are secondary metabolites naturally produced by Aspergillus flavus and Aspergillus parasiticus. These microorganisms usually contaminate grains, nuts and spices both in storage and in the field, and nuts and grains form part of the major animal feed ingredients, including fish feed. Ingestion of aflatoxins has adverse effects on the health of both humans and animals. Inhibiting the growth of these toxigenic Aspergillus spp. will also inhibit the production of aflatoxins. In this study, the effect of coculturing Aspergillus spp. with Pleurotus ostreatus and Saccharomyces cerevisiae on aflatoxin detoxification in fish feed was investigated. The data obtained revealed that coculturing the Aspergillus spp. with the beneficial microorganisms, either singly or together, reduced the aflatoxin content of compounded fish feed (using analysis of variance [ANOVA] at p < .05). There was a low aflatoxin content of 4.96 µg/mg in fish feed contaminated with A. parasiticus and cocultured with S. cerevisiae and P. ostreatus, in comparison with a high aflatoxin content of 13.56 µg/mg in fish feed contaminated with only A. parasiticus. Also, fish feed contaminated with A. flavus and cocultured with S. cerevisiae and P. ostreatus showed a low aflatoxin content of 11.5 µg/mg compared with 19.97 µg/mg in feed contaminated with A. flavus only. Aflatoxin reduction can be attributed to the production of aflatoxin-degrading enzymes by S. cerevisiae and P. ostreatus, and the adsorption of aflatoxins to the cell wall of S. cerevisiae. The findings suggest that coculturing Aspergillus spp. with the beneficial microorganisms is a technique that may improve aflatoxin detoxification, and its application would be beneficial in the food and feed industries. This can be achieved by producing S. cerevisiae in large quantities from local palm wine and sourcing large quantities of P. ostreatus from local farmers. The industrial application of this technique will consequently have a positive effect on the income of local farmers and on the production of safe foods and feed.
Keywords: Aflatoxin detoxification, Coculture, Pleurotus ostreatus, Saccharomyces cerevisiae.
1. Introduction
Aflatoxins are naturally occurring secondary metabolites usually produced by two fungi, Aspergillus flavus and Aspergillus parasiticus. These fungal strains are found naturally all over the world. The four most significant aflatoxins are B1, B2, G1 and G2. A. flavus strains show great variation in their ability to produce aflatoxins (Kumar et al. 2017). Contamination of feeds with aflatoxins is a global challenge (Mwihia et al. 2018). Toxigenic strains of A. flavus are able to produce aflatoxins B1 and B2, and most strains of A. parasiticus can produce all four toxins. These fungal strains usually infect crops on the field and during storage. Most human exposures to aflatoxins occur from nuts, grains and their derived products (WHO, 2018). Billions of people worldwide, mainly in developing countries, are exposed to chronic levels of aflatoxins through contaminated food, either directly or indirectly from the consumption of products from animals fed with contaminated feed.
Adverse effects in fish fed with aflatoxin‐contaminated feed include liver damage, stunting, immunosuppression, decreased growth performance, increased susceptibility to disease and high mortality leading to increased economic losses. The study in Nyeri County revealed that feed samples were contaminated with aflatoxins and could be responsible for poor growth performance, poor appetite, tumours and mortalities observed in fish (Mwihia et al. 2018). The study by Magouz et al. (2018) also shows that Nile tilapia had poor growth when fed with aflatoxin B1‐contaminated feed at 150 ppb. Consequently, the risk of adverse effects of mycotoxins on human beings is increased (Tsafack Takadong et al. 2020). Concentrations of about 20–120 µg/kg body weight per day over a period of 1–3 weeks are acutely toxic (Pickova et al. 2021). Mafe and Büsselberg (2024) stated that the limit of aflatoxin for human consumption is between 30 and 40 µg/kg; therefore, their presence in food and feed is of immense concern.
The production of aflatoxins by toxigenic fungi is supported by the availability of nutrients in substrates, oxygen, high temperature, and high relative humidity in the field or in storage (Ibitoye et al. 2020). The concentration of aflatoxins and their production in feed have been reduced by several methods, which include the use of a non-toxigenic A. flavus strain (NRRL21882) against the toxigenic A. flavus strain in the field, the use of heat, gamma radiation, nixtamalisation, sodium bisulphite and calcium hydroxide. The disadvantages of these treatments are depreciation of nutrients, changes in the taste, colour and texture of foods, a higher cost of food production and chemical-induced food intoxication. Also, non-toxigenic A. flavus may have the ability to produce other mycotoxins apart from aflatoxins (Pickova et al. 2021).
Biological methods, such as the use of microorganisms and enzymes, have overcome these challenges. Some of the microorganisms employed are lactic acid bacteria, certain yeasts of the genera Candida, Debaryomyces, Saccharomyces and Trichosporon, as well as mushrooms of the genus Pleurotus (Ibitoye et al. 2020; Pickova et al. 2021). Volatile compounds, such as esters and alcohol, produced by Saccharomyces cerevisiae inhibited the growth and aflatoxin production of A. flavus (Natarajan et al. 2022). Lactic acid bacteria (LAB) in the study by Møller COdA et al. (2017) inhibited the growth of A. parasiticus and reduced aflatoxin content by 50% in binding studies. Pleurotus spp. are known as cheap sources of ligninolytic enzymes, such as laccase and manganese peroxidase, which are able to degrade aromatic compounds, including aflatoxin, through the destruction of the lactone ring to produce less toxic or non-toxic compounds (Branà et al. 2020; Guan et al. 2021). Pleurotus ostreatus has been reported to degrade aflatoxin (Branà et al. 2017; Wang et al. 2022). Among the yeasts able to degrade aflatoxin, Saccharomyces cerevisiae has been widely useful owing to certain attributes it possesses as a biocontrol agent in foods. It is known as a safe organism used in food fermentations, the production of alcoholic beverages and biofuel. It is easily cultivated on simple nutrients and does not produce allergenic spores, toxins or antibiotics. The ability of S. cerevisiae to control A. flavus is associated with mannan in its cell wall, which binds strongly to various mycotoxins, and with its ability to produce chitinase enzyme (Marwa et al. 2019).
Co-culturing fungi in research is often used to enhance the beneficial effects of the microorganisms, such as increased enzyme production and aflatoxin degradation. For example, the co-culture of Trichoderma spp. and Pleurotus ostreatus yielded sufficient laccase enzyme to detoxify aflatoxins. Also, co-culturing Trametes versicolor with Aspergillus niger produced more laccase enzymes than culturing the single microorganism (Wang et al. 2022). In view of this research development, this study applied the co-cultivation of the known aflatoxin-degrading fungi S. cerevisiae and P. ostreatus in the detoxification of aflatoxin in compounded fish feed for general purposes. Each fungus was used singly in detoxifying aflatoxin in fish feed, and the effect was compared to fish feed containing both fungi using thin layer chromatography (TLC) to detect the aflatoxin levels. This study is geared towards determining the synergistic effect of S. cerevisiae and P. ostreatus as a novel approach in degrading aflatoxin and in its application in fish feed production.
2. Material and Methods
2.1 Collection of Major Fish Feed Ingredients, Mushroom and Palm Wine
Major fish feed ingredients were purchased from four different fish feed mills well known within the Akure metropolis, in polythene bags. These ingredients included maize, soya cake and groundnut cake. The samples were brought to the laboratory for microbiological analysis.
Pleurotus ostreatus spawn was obtained from a culture collection centre, Forestry Research Institute of Nigeria (FRIN), Oyo State, Ibadan, Nigeria, in a sterile bottle and was taken to the laboratory for culturing.
Fresh palm wine was obtained from four retail outlets within Akure metropolis, Ondo State, Nigeria, in sterile containers and was immediately transported to the laboratory for microbiological analysis. The containers were cleansed with 100% ethanol to render them sterile and then closed. Fresh palm wine was transferred into the sterile bottles and immediately sealed.
2.2 Isolation and Identification of Yeast, Toxigenic Fungi and Pleurotus ostreatus
The yeasts were isolated from palm wine using the pour plate method. Three serial dilutions were carried out by diluting 1 ml of palm wine in 9 ml of sterile distilled water. One millilitre of the first and second dilutions was plated, as these dilutions contained a higher population of the target organism, and cultured for 3 days on yeast extract agar (YEA) (HIMEDIA, India) with chloramphenicol to inhibit bacterial growth at ambient temperature. The colonies were enumerated in cfu/ml. Pure isolates were obtained from the mixed culture by sub-culturing thrice using the streak method on YEA. The yeast isolates were tentatively identified by morphological and biochemical characteristics (Ukponobong et al. 2018; Olaniyi et al. 2019).
Aspergillus flavus and Aspergillus parasiticus were isolated from the fish feed ingredients using the pour plate method on sterilised potato dextrose agar (PDA) (HIMEDIA, India) with chloramphenicol to inhibit bacterial growth at ambient temperature (25°C–27°C) for 3 days. One millilitre of the first and second dilutions was plated and cultured for 3 days. The colonies were counted in sfu/ml. Pure fungal isolates were obtained by repeated streaking and identified by cultural and morphological characteristics (Pitt & Hocking, 1997).
Pleurotus ostreatus spawn was cultured on PDA with chloramphenicol at ambient temperature for 4 days and identified by morphological characteristics of the mycelium, by observing its colour and growth pattern (Khan et al. 2011).
2.3 Identification of Yeast, Toxigenic Fungi and Pleurotus ostreatus using Molecular Methods
The isolates were identified by extracting their genomic DNA using the Quick-DNA™ Fungal Kit (Zymo Research, Catalogue No. D6005). The ITS target region was amplified with the primers ITS-1 5’TCCGTAGGTGAACCTGCGG3’ and ITS-4 5’TCCTCCGCTTATTGATATGC3’. The polymerase chain reaction products were run on an agarose gel. The extracted fragments were sequenced in the forward and reverse directions and purified. The purified fragments were analysed on the ABI 3500xl Genetic Analyzer (Applied Biosystems, Thermo Fisher Scientific) for each reaction for each isolate. DNASTAR was used to analyse the .ab1 files generated by the ABI 3500XL Genetic Analyzer, and results were obtained by a BLAST search (NCBI) (Altschul et al. 1997).
2.4 Quantification of Aflatoxins from A. Flavus and A. Parasiticus Biomass
The toxigenic strains of A. flavus and A. parasiticus were sub-cultured on PDA with chloramphenicol (250 mg/200 ml) added to the medium to inhibit bacterial growth, and incubated at ambient temperature for 3 days. Aflatoxins in the fungal biomass were quantified using thin layer chromatography (TLC) in accordance with Leszczynska et al. (2001).
Fifty grammes of fish feed were weighed and transferred into a 250-ml conical flask; 10–15 ml of distilled water and about 200 ml of chloroform were added, and the mouth was plugged with cotton. The flask was mechanically shaken for 1 hour. The sample was filtered and the filtrate was concentrated.
Approximately 30 g of silica gel G (with CaSO₄ as binder) was mixed with 60–65 ml of distilled water in a beaker for about 1 minute. It was then transferred to the applicator and spread uniformly on the plate. The plates were air-dried for 5–10 minutes in dust-free conditions. The gel was activated before use in an oven at 110°C for about 5 minutes. The gel was divided into lanes using a lead pencil. Different known volumes (5 and 10 µl) of the sample extract were spotted in various lanes with a microsyringe on an imaginary line 2.5 cm apart. Similarly, a standard aflatoxin (B1, B2) mixture was spotted in the concentration range of 0.0025 µg in parallel lanes. The plates were developed in a solvent system of toluene : ethyl acetate : formic acid (6 : 3 : 1) in a chromatographic tank for about 50 minutes.
The disintegrated sample was extracted with 10 ml of a methanol–water mixture (7 : 3) to separate aflatoxin. Then, the remainder was homogenised for 10 minutes at room temperature, and the resultant deposit was centrifuged. An aliquot (100 μl) of the supernatant was diluted with 600 μl of phosphate buffer (pH 7.2). The samples were incubated for 30 minutes at room temperature in the dark. Then, 50 μl of tetramethylbenzidine and 50 μl of urea peroxide were added and incubated again for 30 minutes in darkness. The reaction was terminated by adding 100 μl of the stop reagent. The absorbance of the solution was measured at a wavelength of 450 nm using a UV/Visible spectrophotometer. The content of aflatoxins was calculated using a prepared standard curve (Leszczynska et al. 2001).
2.5 Screening of Pleurotus ostreatus and S. Cerevisiae for Aflatoxin-Degrading Enzymes
Laccase activity was determined using a UV–vis spectrophotometer with guaiacol as the substrate. The reaction mixture consisted of 3 ml of 100 mM guaiacol dissolved in 10% acetone in sodium acetate buffer and 1 ml of crude laccase (obtained from a 20-day culture filtrate of P. ostreatus and a 3-day culture filtrate of S. cerevisiae, respectively). The mixture was incubated for 15 minutes, and the absorbance was measured at 470 nm. One unit of laccase activity was determined as the amount of enzyme catalysing the substrate to produce 1 ml of coloured product per minute per ml (Senthivelan et al. 2019).
Manganese peroxidase activity was detected spectrophotometrically. The reaction mixture was prepared by adding 0.1 ml of 0.25 M sodium lactate, 0.05 ml of 2 mM MnSO₄, 0.2 ml of 0.5% bovine serum albumin, 0.1 ml of 0.1% phenol red, 0.5 ml of crude enzyme and 0.05 ml of 2 mM H₂O₂ in 0.2 M sodium phosphate buffer (pH 8.0). The mixture was kept at ambient temperature for 5 minutes and the reaction was stopped with 0.04 ml of 2 N NaOH. The absorbance was measured at 610 nm. One activity unit was defined as the amount of enzyme necessary to oxidise 1 µmol of substrate per minute (Patricia Lopes et al. 2009).
Chitinase activity was measured using colloidal chitin as the substrate. One millilitre of a 3-day yeast broth (enzyme solution) was allowed to react with 1 ml of 0.5% colloidal chitin in 0.1 M citrate buffer (pH 7.0). The mixture was incubated at 37°C in a shaking water bath for 30 minutes. The reaction was stopped by adding 2 ml of DNS reagent after incubation. Then, the mixture was heated in a boiling water bath for 5 minutes and allowed to cool. The cooled solution was centrifuged at 10,000 rpm at room temperature for 5 minutes. The absorbance of the supernatant was measured at 540 nm against the control. The supernatant was used for the analysis of reducing sugar using the dinitrosalicylic acid (DNS) method. One unit of chitinase activity was determined as the amount of enzyme that liberates 1 μmol of reducing sugar per minute per ml (Gonfa et al. 2023).
2.6 Antifungal Activity of Pleurotus ostreatus and S. Cerevisiae against the Toxigenic Fungi
The antifungal activity of P. ostreatus and S. cerevisiae against toxigenic A. flavus and A. parasiticus was carried out according to Marwa et al. (2019). Yeast extract agar plates were inoculated with 0.5 ml of 2-day-old spore suspensions of A. flavus and A. parasiticus (108 sfu/ml, respectively), and 20 µl of a 24-hour cell suspension of S. cerevisiae (106 cells/ml) and a 17-day cell suspension of Pleurotus ostreatus (0.38 mycelium turbidity at 640 nm) were spotted in the centre of each plate (6-mm hole diameter). The inoculated plates were incubated for 8 days, and the zone of inhibition of the toxigenic fungi was recorded every 24 hours using the agar well diffusion method.
2.7 Formulation of Fish Feed using the Major Fish Feed Ingredients
Fish feed was formulated using the Pearson square method to obtain 40% crude protein. The fish feed formulation included the major ingredients: soya cake, groundnut cake, maize, fish meal, limestone, salt, bonemeal, binder, vegetable oil, vitamin premix, mineral premix and wheat offal (Arsenos et al. 2023).
2.8 Detoxification of Artificially Contaminated Formulated Fish Feed Using P. ostreatus and S. Cerevisiae
Four hundred grammes (400 g) of the formulated fish feed was divided into eight portions and sterilised in a laboratory oven at 110°C for 2 hours. The sterilised samples were allowed to cool and were then inoculated with or without fungal spore suspension. Before inoculation, 15 ml of sterile distilled water was added to moisten the samples to enhance infectivity, followed by inoculation with the toxigenic fungus and incubation for 4 days at ambient temperature. After this, the samples were inoculated again with S. cerevisiae (Y) for another 4 days or with P. ostreatus (M) for another 4 days, according to Ibitoye et al. (2020) with slight modification. The first portion contained the formulated fish feed inoculated with 0.4 ml spore suspension of A. parasiticus (MF) (control), while the second portion contained fish feed inoculated with 0.4 ml spore suspension of MF plus 0.4 ml of Y cell suspension (MFY). The third portion contained fish feed inoculated with 0.4 ml spore suspension of MF plus 0.4 ml mycelium suspension of M (MFM); the fourth portion contained fish feed inoculated with 0.4 ml spore suspension of MF plus 0.4 ml of Y plus 0.4 ml mycelium suspension of M (MFMY). The fifth portion contained the formulated fish feed inoculated with 0.4 ml spore suspension of A. flavus (SP) (control), while the sixth portion contained fish feed inoculated with 0.4 ml spore suspension of SP plus 0.4 ml of Y cell suspension (SPY). The seventh portion contained fish feed inoculated with 0.4 ml spore suspension of SP plus 0.4 ml mycelium suspension of M (SPM); the eighth portion contained fish feed inoculated with 0.4 ml spore suspension of SP plus 0.4 ml of Y plus 0.4 ml mycelium suspension of M (SPMY). After detoxification, each portion was oven-dried at 70°C for 1 hour. This step was performed to terminate the in vitro antifungal reactions in the formulated fish feed. The aflatoxin content in each sample was thereafter quantified (Ibitoye et al. 2020).
2.9 Statistical Analysis
Analysis of variance (ANOVA) at p < .05 was used to analyse the data using IBM SPSS Statistics version 23. Significant differences in the aflatoxin content of the formulated fish feeds were compared using post hoc tests and Duncan’s test for the separation of means.
3. Results
3.1 Total Yeast and Fungal Counts
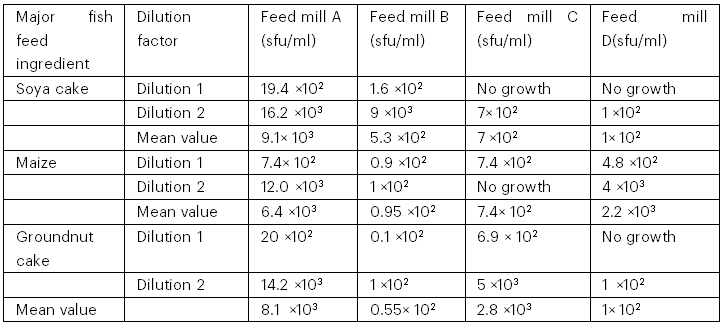
The total yeast counts from palm wine are presented in Table 1 and the total fungal counts are presented in Table 2. The palm wine sample from Ibule Local Government Area had the highest yeast count of 13.1 × 10³ cfu/ml, while the palm wine sample from Oba-Ile had the lowest yeast count of 7.9 × 10³ cfu/ml. The fungal counts from the major fish feed ingredients revealed that feed mill A had the highest fungal count in soya cake (9.1 × 10³ sfu/ml), while feed mill D had the lowest fungal count (1 × 10² sfu/ml). Feed mill A had the highest fungal counts in maize (6.4 × 10³ sfu/ml), while feed mill B had the lowest fungal count (0.95 × 10² sfu/ml); in groundnut cake, feed mill A had the highest fungal counts (8.1 × 10³ sfu/ml) while feed mill B had the lowest count (0.55 × 10² sfu/ml).
Table 1: Total Yeast Counts from Palm Wine Obtained from Four Different Outlets

Table 2: Total Fungal Counts from Major Fish Feed Ingredients Obtained from Four Feed Mills
3.2 Morphological and Biochemical Characteristics of Isolated Yeasts
Tables 3 and 4 show the morphological and biochemical characteristics of the isolated yeasts. The yeasts were suspected to be Candida spp. and Saccharomyces spp. based on morphological and biochemical tests.
Table 3: Colonial and Microscopic Characteristics of Isolated Yeasts

Table 4: Biochemical Test Results of the Isolated Yeasts
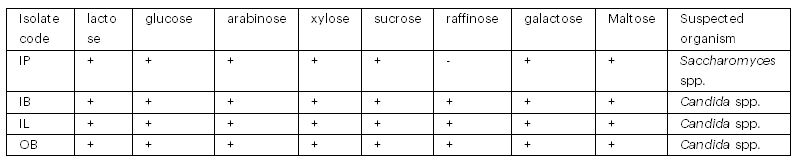
Key: IP- Ipinsa, IB- Ibule, IL- Ilara, OB- Oba-Ile
3.3 Morphological Characteristics of Isolated Fungi and Pleurotus ostreatus
Table 5 reveals the morphological features of the isolated fungi. The fungal isolates were suspected to be Aspergillus parasiticus and Aspergillus flavus using cultural and microscopic characteristics. The P. ostreatus mycelium was white, cotton-like, tufted and fluffy, according to Khan et al. (2011).

3.4 Molecular Identity of the Yeast, Toxigenic Fungi and Pleurotus ostreatus
The suspected organisms were given codes and molecular identification was carried out. The yeast that had antifungal activity against the toxigenic fungi was identified using molecular techniques. The BLAST prediction of GenBank for all the isolates gave percentage identities between the fungi tested and those detected in GenBank, as shown in Table 6. The ITS rDNA sequence analysis identified the isolates as Aspergillus flavus, Aspergillus parasiticus, Pleurotus ostreatus and Saccharomyces cerevisiae.
Table 6: Molecular Identity of the Yeast, Toxigenic Fungi and Pleurotus ostreatus

3.5 Aflatoxins Contents in Toxigenic A. Flavus and A. Parasiticus
The isolated A. flavus and A. parasiticus biomass produced aflatoxins that were quantified as 1.5630 µg/mg and 0.4141 µg/mg, respectively (Table 7).
Table 7: Aflatoxins Contents in Toxigenic A. flavus and A. parasiticus Biomass

3.6 Aflatoxin-Degrading Enzyme Activity in P. ostreatus and S. Cerevisiae
The cultured P. ostreatus and isolated S. cerevisiae were screened for the production of aflatoxin-degrading enzymes. Table 8 shows that P. ostreatus produced laccase and manganese peroxidase enzymes, while S. cerevisiae produced laccase and chitinase enzymes.
Table 8: Aflatoxin-Degrading Enzyme Activity in P. ostreatus and S. cerevisiae

3.7 Antifungal Activity of P. ostreatus and S. Cerevisiae against Toxigenic A. Flavus and A. Parasiticus
Table 9 shows the antifungal growth inhibitory potency of P. ostreatus and S. cerevisiae against toxigenic A. flavus and A. parasiticus. The zone of inhibition was best on the second day for both P. ostreatus and S. cerevisiae. The highest zone of inhibition was recorded for S. cerevisiae against A. parasiticus on the second day, while the lowest zone of inhibition was recorded for P. ostreatus against A. parasiticus on the first day. The zone of inhibition remained constant from 72 hours until the eighth day.
Table 9: Antifungal Potentials of P. ostreatus and S. cerevisiae Against Toxigenic A. flavus and A. parasiticus
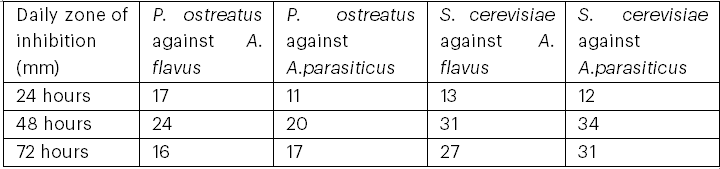
The zone of inhibition of S. cerevisiae against the toxigenic fungi was wider in comparison to that of P. ostreatus, suggesting greater effectiveness of S. cerevisiae against toxigenic fungi (Bayston, 2017).
3.8 Aflatoxin Content of Contaminated and Treated Fish Feed Samples
Figure 1 shows the quantities of aflatoxins in A. parasiticus-contaminated and treated compounded fish feed samples. The A. parasiticus-contaminated feed had the highest aflatoxin content (13.56 µg/mg), followed by the contaminated feed treated with P. ostreatus (11.38 µg/mg), then the contaminated feed treated with S. cerevisiae (10.53 µg/mg), and finally the contaminated feed treated with the co-culture of P. ostreatus and S. cerevisiae had the lowest aflatoxin content (4.96 µg/mg).
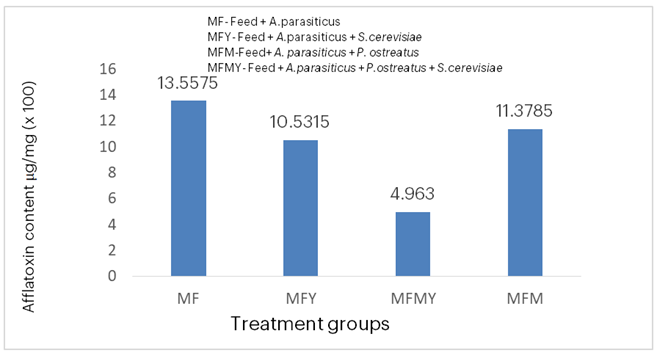
Figure 1: Aflatoxin Quantities in A. parasiticus-Contaminated and Treated Compounded Fish Feed.
Figure 2 shows that the A. flavus-contaminated fish feed had the highest aflatoxin content (19.97 µg/mg), followed by the contaminated feed treated with P. ostreatus (18.04 µg/mg). However, the contaminated feed treated with the co-culture of P. ostreatus and S. cerevisiae had a lower aflatoxin content (11.5 µg/mg) compared with the contaminated feed, and the A. flavus-contaminated feed treated with S. cerevisiae had the lowest aflatoxin content (9.68 µg/mg).

Figure 2: Aflatoxin Quantities in A. flavus-Contaminated and Treated Compounded Fish Feed.
4. Discussion
Food, typically meat, can be spoiled by Clostridium species which results in gas and foul odour (Kyrylenko et al., 2023). The poor sanitary habits of meat handlers may be as a result of low level of education. Gashe et al. (2020) is in alignment with the study where he reported that the meat handlers’ limited understanding of food handling and processing procedures is due to their low level of education Food handlers can be trained in fundamental principles and personal hygiene to ensure food safety (Bersia et al., 2020). Just 18% (2) of the food workers in this sample developed skin problems.
Varying fungal counts were obtained from the major fish feed ingredients from the four different feed mills, and toxigenic fungi, Aspergillus flavus and Aspergillus parasiticus, were found among the fungal species. This is in agreement with the findings of Guan et al. (2021), who stated that cereals—one of the major ingredients of feeds—are the target of toxigenic fungi, which contaminate feed and produce aflatoxins. Also, Shabeer et al. (2022) reported that A. flavus is a common contaminant of feed. This is similar to the result of this study, where A. flavus was isolated from the fish feed ingredients used in fish feed formulation.
Different yeast counts were obtained from palm wine from four different local government areas, with Ibule LG having the highest yeast count (13.1 × 10³), followed by Ilara LG (10.7 × 10³), Ipinsa LG (10.2 × 10³) and Oba-Ile LG (7.9 × 10³). The suspected yeasts were Candida spp. and Saccharomyces spp. They were identified using colonial and microscopic methods to detect their colours, elevations and shapes, respectively. Biochemical tests were used to determine the sugars they could ferment. These findings support Djeni et al. (2022), who reported that fresh palm wine contains different microorganisms, such as yeasts and bacteria, and that it is dominated by S. cerevisiae, as the yeasts isolated in this study were obtained from freshly tapped palm wine. Olaniyi et al. (2019) also reported that varying yeast counts might be due to different sources, times of collection, environmental conditions and storage containers. This is in agreement with the results obtained in this study, as Ibule LG palm wine—with the highest yeast count—was sourced from raffia palm trees, compared with other local government areas where palm wine was tapped from oil palm trees. They also reported that palm wine is rich in sugars that can be fermented by various yeasts.
The 72-hour study of the antifungal activity of P. ostreatus against A. flavus and A. parasiticus produced optimal zones of inhibition (24 mm and 20 mm, respectively) at 48 hours. This suggests that the toxigenic fungi resuscitated after 48 hours, as there was no continuous application of the broth containing the antifungal compounds. This is in agreement with Hussain et al. (2020), who observed A. flavus inhibition using Pleurotus spp. extract, although the extracts in that study were obtained using 95% methanol. The best zones of inhibition produced in the study of the antifungal activity of S. cerevisiae against A. flavus and A. parasiticus were 31 mm and 34 mm, respectively, at 48 hours. This is in agreement with Yang et al. (2020), who reported that S. cerevisiae inhibited the growth of A. flavus due to the production of volatile compounds—including 3-methyl-1-butanol—that have growth inhibitory activities. Marwa et al. (2019) also stated that S. cerevisiae inhibited the growth of A. flavus, which can be associated with competition for nutrients and the synthesis of hydrolytic enzymes. According to Freimoser et al. (2019), one of the underlying biocontrol mechanisms of S. cerevisiae is the secretion of hydrolytic enzymes, such as chitinase.
The co-cultivation of a beneficial microorganism with a toxin-producing microorganism reduced toxin production due to enhanced production of aflatoxin-degrading enzymes. These enzymes were able to destroy the lactone rings of aflatoxin (Wang et al. 2022). In this study, the co-cultivation of S. cerevisiae and P. ostreatus—used singly and together with toxigenic A. flavus and A. parasiticus, respectively—resulted in a significant reduction of aflatoxins in compounded fish feed. There were reductions in aflatoxin contents to 4.96, 10.53 and 11.38 µg/mg in fish feed contaminated with A. parasiticus cocultured with both S. cerevisiae and P. ostreatus, with S. cerevisiae only, and with P. ostreatus only, respectively, compared to the high aflatoxin content of 13.56 µg/mg in fish feed contaminated with A. parasiticus only. Also, there were reductions in aflatoxin contents to 9.68, 11.5 and 18.03 µg/mg in fish feed contaminated with A. flavus cocultured with S. cerevisiae only, with both S. cerevisiae and P. ostreatus, and with P. ostreatus only, respectively, compared to the high aflatoxin content of 19.97 µg/mg in fish feed contaminated with A. flavus only. Similar results were obtained in the co-culturing of A. flavus and A. parasiticus with Salmonella, in which there was a reduction in the quantities of aflatoxins B1, B2, G1 and G2 produced by these fungi (Von Hertwig et al. 2020). Ibitoye et al. (2020) reported a similar finding, where co-culturing A. flavus with lactic acid bacteria in millet resulted in a low aflatoxin content of 6.9 µg/kg compared to millet contaminated with A. flavus only (15.25 µg/kg). Also, co-culturing a non-toxigenic strain of A. flavus reduced the aflatoxin content in maize grown for feed production in the field. The success of co-culturing can be linked to competition that excludes and inhibits the growth of the toxin-producing or pathogenic microorganism (Sarocco et al. 2019). This is in agreement with the antifungal activity results of P. ostreatus and S. cerevisiae in this study, where the growth of toxigenic A. flavus and A. parasiticus was inhibited using the agar well diffusion method. Clustering of genes or mutations may also contribute to the effectiveness of co-culturing (Sarocco et al. 2019). The pathway that produces aflatoxins is also disrupted; according to Caceres et al. (2018), the combined cultivation of A. flavus with Streptomyces roseolus reduced aflatoxin production. This finding suggests that the pathway of aflatoxin production was disrupted in this study, as aflatoxin contents were reduced in fish feed contaminated with Aspergillus spp. cocultured with either S. cerevisiae, P. ostreatus or a combination of both. Co-cultivation suppresses the growth and aflatoxin production by A. flavus by inhibiting the expression of aflatoxin-promoting regulators and increasing the expression of aflatoxin-inhibiting regulators (Yang et al. 2020). The data from this study revealed that reducing aflatoxin content in fish feed using monocultures of S. cerevisiae with either toxigenic A. flavus or A. parasiticus was more effective than using monocultures of P. ostreatus with either fungus. This can be attributed to the ability of Saccharomyces spp. to strongly adsorb aflatoxins to their cell wall via van der Waals forces, forming a complementary structure that does not easily dissociate. This characteristic makes Saccharomyces spp. useful in food detoxification (Guan et al. 2021). However, certain physicochemical conditions, such as pH and temperature, determine effective toxin adsorption by microorganisms (Kowalczyk et al. 2021); therefore, further research is needed to optimise conditions that improve the adsorbing ability of S. cerevisiae and the co-culturing technique to further reduce aflatoxins. According to Guan et al. (2021), it is advantageous to use probiotics in the detoxification process as they exert a dual effect, including the adsorption of aflatoxins and their conversion to less toxic compounds. In this study, the co-culturing of S. cerevisiae and P. ostreatus with A. parasiticus greatly reduced the aflatoxin content in compounded fish feed from 13.55 µg/mg in the contaminated feed to 4.96 µg/mg, which was the most effective reduction observed among the co-culturing treatments. This might be due to S. cerevisiae being a probiotic and P. ostreatus being an edible fungus that produces aflatoxin-degrading enzymes. The simple method of mixing the cell suspensions of S. cerevisiae and P. ostreatus with fish feed might be applied as a pretreatment to reduce aflatoxin content before the feed is administered to fish.
5. Conclusion
Findings from this study establish the ability of P. ostreatus and S. cerevisiae to inhibit the growth of toxigenic A. flavus and A. parasiticus. These microorganisms, which are regarded as probiotics, will serve as safe, cheap and eco-friendly agents for inhibiting aflatoxin production in fish feed, thereby producing safe and nutritious feed devoid of chemical residues. Further research is required to gain a better understanding of the mechanisms behind co-culturing, which will determine the proper dosage of the microorganisms and the optimal conditions for improved aflatoxin reduction.
Acknowledgement
The authors are grateful to the Department of Microbiology, School of Life Sciences, The Federal University of Technology, Akure, Ondo State, Nigeria, for providing the necessary support in terms of equipment, technical advice during the experimental procedure, and laboratory facilities used for the study. The authors are also grateful to the Forestry Research Institute of Nigeria (FRIN) for providing the spawns of P. ostreatus and to Iquaba Biotech for the molecular identification of the microorganisms.
Funding
The authors declare that no funds, grants or other support were received during the preparation of this manuscript. Lack of funding does not influence the scope of the study.
Author’s Contributions
CrediT author statement
Akinyele, B. J.: Conceptualisation, Methodology; Olalemi, O. A.: Supervision, Writing—Reviewing and Editing; Oluwole, O. R.: Data Curation, Writing—Original Draft Preparation.
Conflict of Interest
The authors have no relevant financial or non-financial interests to disclose.
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Research, 25(17), 3389–3402.
Bayston, R. (2017). Comprehensive biomaterials II. Elsevier.
Branà, M. T., Cimmarusti, M. T., Haidukowski, M., Logrieco, A. F., & Altimare, C. (2017). Bioremediation of aflatoxin B₁‐contaminated maize by king oyster mushroom (Pleurotus eryngii). PLOS ONE. https://doi.org/10.1371/journa l.pone.0182574
Branà, M. T., Sergio, L., Haidukowski, M., Logrieco, A. F., & Altomare, C. (2020). Degradation of aflatoxin B₁ by a sustainable enzymatic extract from spent mushroom substrate of Pleurotus eryngii. Toxins, 12(1), 49.
Caceres, I., Snini, S. P., Puel, O., & Mathieu, F. (2018). Streptomyces roseolus, a promising biocontrol agent against Aspergillus flavus, the main aflatoxin B₁ producer. Toxins, 10, 442.
Djeni, T. N., Keisam, S., Kouame, K. H., Assohoun-Djeni, C. N., Ake, F. D., Amoikon, L. S., & Jeyaram, K. (2022). Dynamics of microbial populations and metabolites of fermenting saps throughout the tapping process of ron and oil palm trees in Côte d’Ivoire. Frontiers in Microbiology, 13, 954917.
Gonfa, T. G., Negessa, A. K., & Bulto, A. O. (2023). Isolation, screening and identification of chitinase‐producing bacterial strains from riverbank soils at Ambo, Western Ethiopia. Heliyon, 9(11), e21643.
Guan, Y., Chen, J., Nepovimova, E., Long, M., Wu, W., & Kuca, K. (2021). Aflatoxin detoxification using microorganisms and enzymes. Toxins, 13(1), 46. https://do i.org/10.3390/toxins13010046
Hussain, A. H., & Hussein, H. Z. (2020). Evaluation of Agaricus sp. and Pleurotus sp. extracts efficiency in Aspergillus flavus growth inhibition and aflatoxin B₁ reduction. Systematic Reviews in Pharmacy, 11(10), 564–569.
Ibitoye, O. A., Olaniyi, O. O., Ogidi, C. O., & Akinyele, B. J. (2020). Lactic acid bacteria bio‐detoxified aflatoxins‐contaminated cereals, ameliorate toxicological effects and improve haemato‐histological parameters in albino rats. Toxin Reviews. Advance online publication. https://doi.org/10.1080 /15569543.2020.1817088
Kowalczyk, P., Ligas, B., Skrzypczak, D., Mikula, K., Izydorczyk, G., Witek-Krowiak, A., Moustakas, K., & Chojnacka, K. (2021). Biosorption as a method of biowaste valorisation to feed additives: RSM optimisation. Environmental Pollution, 268, 115937.
Khan, S. M., Nawaz, A., Malik, W., Javed, N., Yasmin, T., ur Rehman, M., & Khan, A. A. (2011). Morphological and molecular characterisation of oyster mushroom (Pleurotus spp.). African Journal of Biotechnology, 10(14), 2638–2643.
Kumar, P., Mahato, D. K., Kamle, M., Mohanta, T. K., & Kang, S. G. (2017). Aflatoxins: A global concern for food safety, human health and their management. Frontiers in Microbiology, 7, 2170. https://doi.org/10.3389/fmicb.2016.02170
Leszczynska, J., Owczarek, A., & Maslowska, U. (2001). Determination of aflatoxin in food products. Czech Journal of Food Sciences, 19, 8–12.
Mafe, A. N., & Büsselberg, D. (2024). Mycotoxins in food: Cancer risks and strategies for control. Foods, 13(21), 3502.
Magouz, F. I., Salem, M. S., & Hashad, M. A. (2018). Effect of some mycotoxins on growth performance and feed utilisation of Nile tilapia (Oreochromis niloticus). Iraqi Journal of Veterinary Sciences, 32(1).
Marwa, A. M., Ramsey, A. M., & Zohri, A. A. (2019). The action mechanism and biocontrol potentiality of novel isolates of Saccharomyces cerevisiae against the aflatoxigenic Aspergillus flavus. Letters in Applied Microbiology, 68(2), 104–111.
Moller, C. O. D. A., Freire, L., Rosim, R. E., Margalho, L. P., Balthazar, C. F., Franco, L. T., & Bayston, R. (2017). Comprehensive biomaterials II. Elsevier.
Natarajan, S., Balachandar, D., Senthil, N., Velazhahan, R., & Paranidharan, V. (2022). Volatiles of antagonistic soil yeasts inhibit growth and aflatoxin production of Aspergillus flavus. Microbiological Research, 263, 127150.
Olaniyi, O. O., Akinlami, O. R., Adeleke, B. S., Alabi, G. O., & Akinyele, B. J. (2019). Isolation and characterisation of alcohol-tolerant Saccharomyces cerevisiae from palm wine (Raffia palm). Annals of Food Science and Technology, 20(4), 701–709.
Patricia Lopes, O., Marta Cristina, T. D., Alexandre, N. P., & Lucia, R. D. (2009). Purification and partial characterisation of manganese peroxidase from Bacillus pumilus and Paenibacillus sp. Brazilian Journal of Microbiology, 40, 818–826.
Pickova, D., Ostry, V., Toman, J., & Malir, F. (2021). Aflatoxins: History, significant milestones, recent data on their toxicity and ways to mitigation. Toxins, 13, 399.
Pitt, J. I., & Hocking, A. D. (2022). Ecology of fungal food spoilage. In Fungi and food spoilage (pp. 3–12). Springer International Publishing.
Senthivelan, T., Kanagaraj, J., Panda, R. C., & Narayani, T. (2019). Screening and production of a potential extracellular fungal laccase from Penicillium chrysogenum: Media optimisation by response surface methodology (RSM) and central composite rotatable design (CCRD). Biotechnology Reports, 23, e00344.
Shabeer, S., Jamal, A., & Ali, A. (2022). Aflatoxin contamination, its impact and management strategies: An updated review. Toxins, 14(5), 307. https://doi.org/10.3390/toxins14050307
Tsafack Takadong, J. J., Mouafo, H. T., Manet, L., Baomog, A. M. B., Adjele, J. J. B., Medjo, E. K., & Medoua, G. N. (2020). Assessment of the presence of total aflatoxins and aflatoxin B₁ in fish fermented in two Cameroonian localities. International Journal of Food Science, Article ID 2506812.
von Hertwig, A. M., Iamanaka, B. T., Neto, D. P. A., de Rezende, J. B., Martins, L. M., Taniwaki, M. H., & Nascimento, M. S. (2020). Interaction of Aspergillus flavus and A. parasiticus with Salmonella spp. isolated from peanuts. International Journal of Food Microbiology, 328, 108666.
Wang, L., Huang, W., Shen, Y., Zhao, Y., Wu, D., Yin, H., & Wang, J. (2022). Enhancing the degradation of aflatoxin B₁ by co-cultivation of two fungi strains with the improved production of detoxifying enzymes. Food Chemistry, 371, 131092.
World Health Organization. (2018). Food Safety Digest: Aflatoxins (REF No: WHO/NMH/FOS/RAM/18.1).
Yang, K., Geng, Q., Song, F., He, X., Hu, T., Wang, S., & Tian, J. (2020). Transcriptome sequencing revealed an inhibitory mechanism of Aspergillus flavus asexual development and aflatoxin metabolism by soy-fermenting non-aflatoxigenic Aspergillus. International Journal of Molecular Sciences, 21, 6994.
About this Article
Cite this Article
APA
Oluwole, O. R., Olalemi, A. O. & Akinyele, B. J. (2025). Coculture of Saccharomyces cerevisiae and Pleurotus ostreatus Improves Aflatoxin Detoxification in Fish Feed. In Akinyele B.J., Kayode R. & Akinsemolu A.A. (Eds.), Microbes, Mentorship, and Beyond: A Festschrift in Honour of Professor F.A. Akinyosoye. SustainE
Chicago
Oluwole, O. R., Olalemi, A. O. and Akinyele, B. J. 2025. “Coculture of Saccharomyces cerevisiae and Pleurotus ostreatus Improves Aflatoxin Detoxification in Fish Feed.” In Microbes, Mentorship, and Beyond: A Festschrift in Honour of Professor F.A. Akinyosoye, edited by Akinyele B.J., Kayode R. and Akinsemolu A.A., SustainE.
Received
1 November 2024
Accepted
15 January 2025
Published
4 February 2025
Corresponding Author Email: omowunmi.oluwole@gmail.com
Disclaimer: The opinions and statements expressed in this article are the authors’ sole responsibility and do not necessarily reflect the viewpoints of their affiliated organizations, the publisher, the hosted journal, the editors, or the reviewers. Furthermore, any product evaluated in this article or claims made by its manufacturer are not guaranteed or endorsed by the publisher.
Distributed under Creative Commons CC-BY 4.0
Share this article
Use the buttons below to share the article on desired platforms.














