Eniola K.I.T. 1 David O.M. 2 Ajayi P.O. 3 Ajayi O.O. 4 & Adeyemo-Eleyode V.O. 5
1Environmental and Public Health Research Laboratory, Department of Biological Sciences, Joseph Ayo Babalola University, Ikeji Arakeji, Osun State, Nigeria.
2Department of Microbiology, Ekiti State University, Ado Ekiti, Ekiti State, Nigeria.
*Corresponding Author Email: kennyeniola@gmail.com …
Abstract
Managing bacterial infections becomes particularly challenging when antibiotic-resistant organisms are involved, especially multiple antibiotic-resistant (MAR) strains. Leafy vegetables consumed raw can harbor MAR pathogens, serving as transmission vehicles to consumers. This study examined the co-presence of antibiotic resistance and virulence genes in MAR strains among 18 Escherichia coli isolates from retailed leafy vegetables. Eight strains with MAR index > 0.6 were selected and screened for resistance genes (parC, aad2, TEM, qnrA, aac(3)-IV, shv, ctx-m, ant(2″)-I) and virulence genes (fimH, usp, stx1). Casein, gelatinase, haemolysis, and proteinase activities were also assessed. The TEM gene was detected in all isolates, while shv, aad2, ctx-m, ant(2″)-I, qnrA, and aac(3)-IV were found in varying frequencies. Among virulence genes, usp was present in seven isolates, while stx1 and fimH were detected in five. Two isolates carried all three virulence genes. Eight distinct co-presence patterns of resistance and virulence genes were identified. These findings highlight potential challenges in managing infections caused by such isolates, emphasizing the need for robust antibiotic stewardship and careful handling of leafy vegetables, particularly those consumed raw.
Keywords: Antibiotic resistance, multiple antibiotic-resistant (MAR) strains, Escherichia coli, virulence genes, foodborne transmission.
1. Introduction
Diarrhoeal diseases are associated with high morbidity and mortality rates worldwide, particularly in developing countries. These high rates are primarily attributable to poor sanitation and hygiene, lack of safe drinking water supplies, open defaecation, and the use of contaminated water sources for farming, cooking, and other activities (Swedan & Alrub, 2019; Natvig et al., 2002). In developing countries, diarrhoeal diseases are the second leading cause of death in children under the age of five (following pneumonia) (UNICEF, 2009) and have emerged as a leading cause of childhood mortality in recent years (UNICEF, 2024).
Many pathogens are implicated in diarrhoeal diseases, including pathogenic strains of E. coli. These strains are classified into six pathotypes that are collectively referred to as diarrhoeagenic E. coli. E. coli is a leading foodborne pathogen and has been responsible for several outbreaks (CDC, 2014). According to a report by the CDC (2020), there were at least three separate outbreaks of E. coli infections in the United States between June and December 2020; one of these outbreaks was linked to leafy vegetables, resulting in 90 reported infections, 41 hospitalisations and one death. A study conducted in fifteen populations across countries with low and medium human development indices from 1984 to 2005 reported incidences ranging from 39 to 460 infections per 1,000 persons per year (Gupta et al., 2008).
Escherichia coli O157:H7 is particularly lethal due to additional genes that enable adhesion to the intestinal wall and the production of the highly virulent Shiga toxin. This toxin damages cells in the intestinal wall and blood vessels, resulting in haemorrhage and renal failure (Rasheed et al., 2014). Furthermore, infection with enterotoxigenic E. coli (ETEC) is a leading cause of traveller’s diarrhoea and a major cause of diarrhoeal disease in underdeveloped nations, especially among children (Ratchtrachenchai et al., 2004). ETEC produces two types of toxins: the heat-labile toxin (LTh) and the heat-stable toxin (STh). Both toxins stimulate the intestinal lining to secrete excessive fluid, leading to profuse watery diarrhoea. Although different ETEC strains may secrete either one or both toxins, the clinical manifestations of the illness are similar. Currently, the identification of ETEC is based on the detection of their virulence markers (CDC, 2014; Rasheed et al., 2014).
Vegetables are excellent sources of vitamins, minerals and dietary fibre (Dias, 2012; Olaimat & Holley, 2012). Increased awareness of the health benefits associated with leafy vegetable consumption has led to a rise in demand over the past two decades. However, the cultivation of vegetables through organic farming has been described as a potential risk to public health owing to the likelihood of pathogen contamination (Tango et al., 2014). Vegetables can become contaminated with foodborne pathogens during several stages, including cultivation, harvesting, postharvest handling, processing and distribution (Beuchat et al., 2008). Bacteria, including pathogens, adhere to vegetables depending on the surface properties of the leaves and stems. Dust and soil present on vegetables or sprouts are typically removed by a simple water wash, which is often insufficient for eliminating bacteria (Van Haute et al., 2015). Additionally, the water used to wash these vegetables may itself be a source of contamination (Rai et al., 2007). Consequently, the consumption of contaminated vegetables—whether directly or in salads and similar preparations—poses significant public health threats. The presence of E. coli on commonly consumed fruits and vegetables has been previously reported (Rasheed et al., 2014).
In this study, we examine the co-presence of antibiotic resistance genes and virulence genes in multiple antibiotic-resistant E. coli strains isolated from leafy vegetables sold in major markets.
2. Materials and Methods
Source of Escherichia coli Strains
The study was conducted using eighteen E. coli strains previously isolated from commonly consumed leafy vegetables, including Amaranthus hybridus (‘Efo tete’), Corchorus olitorius (‘Ewedu’), Crassocephalum crepidioides (‘Ebolo’), Solanecio biafrae (‘Worowo’), Talinum triangulare (‘Gure’), and Telfairia occidentalis (‘Ugu’). These vegetables were retailed in the two major markets around Joseph Ayo Babalola University, Ikeji Arakeji (7.36°N, 5.10°E), as reported in a previous study (Eniola et al., 2021).
2.2 Screening of Isolates for Multiple Antibiotic Resistance
Pure broth cultures of the isolates were prepared and standardised to 0.5 McFarland, following the method described by Eniola et al. (2021). The isolates were then screened for resistance to various antibiotics using the Kirby–Bauer disk diffusion method on Mueller–Hinton agar (Lim et al., 2007). The susceptibility and/or resistance of the E. coli isolates were determined in accordance with the Clinical Laboratory Standards Institute (2012) guidelines, and the antibiotic resistance profiles were established. The multiple antibiotic resistance (MAR) index for each isolate was calculated as described by Krumperman (1983), and isolates with a MAR index equal to or greater than 0.75 were selected for further study.
2.3 Screening for Antibiotic Resistance (Ar) Genes and Virulence Genes in the Isolates
Genomic DNA was extracted from the selected MAR E. coli strains, and the DNA was amplified using the polymerase chain reaction (PCR) before purification, following the protocols of Makhado et al. (2022) and Johnning et al. (2015). The genomes were subsequently screened for the presence of antibiotic resistance genes (parC, aad2, TEM, qnrA, aac(3)-IV, shv, ctx-m, and ant(2″)-I) and virulence genes (fimH, usp, and stx1) by PCR amplification using the relevant primers (see Tables 1 and 2).
Table 1: Primers for the Detection of Antibiotic-Resistant Genes in the Isolates
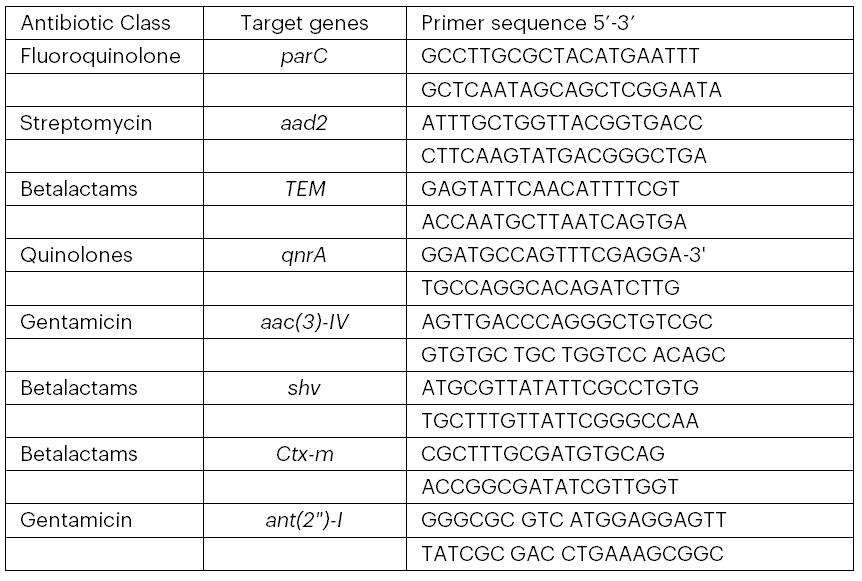
Table 2: Primers Used to Detect the Virulence Genes

2.4 Detection of Virulence Factors in the Isolates
The selected MAR isolates were further screened for specific virulence factors. Casein activity was determined according to the modified methods described by Grant et al. (2008) and Haddadi et al. (2005). Gelatinase, haemolysis and proteinase activities were assessed using the procedures outlined by Gupta and Malik (2007).
3. Results
3.1 Antibiotic Susceptibility of Escherichia Coli Isolates from Leafy Vegetables
TAll eighteen E. coli isolates were resistant to at least two antibiotics, with two isolates exhibiting resistance to all the antibiotics tested. Thirteen distinct antibiotic resistance profiles were observed among the isolates (see Table 3). The most common resistance patterns were resistance to two and to six antibiotics, with five isolates each displaying these profiles. The multiple antibiotic resistance (MAR) index ranged from 0.25 (in five isolates) to 1.00 (in two isolates), with eight isolates having a MAR index equal to or greater than 0.75 (see Table 3).
3.2 Presence of Antibiotic Resistance Genes
Antibiotic resistance genes were detected in the eight MAR isolates selected for further analysis. These isolates carried between four and seven antibiotic resistance (AR) genes (see Table 4). The TEM gene was detected in all eight isolates, whereas the aac(3)-IV gene was present in only two isolates. None of the isolates harboured every AR gene assayed (see Plates 1–8).
3.3 Presence of Virulence Genes
Virulence genes fimH, usp and stx1 were detected in the eight MAR E. coli strains examined. The isolates carried between one and three virulence genes. All the virulence genes assayed were present in only three isolates. Specifically, the fimH gene was detected in five isolates, the usp gene in seven isolates, and the stx1 gene in five isolates (see Table 5).
3.4 Presence of Virulence Factors in Mar E. coli Strains from Leafy Vegetables
Screening for enzymatic virulence factors revealed that such factors were detected in only four of the isolates. In one isolate, three factors—β-haemolytic activity, caseinase and gelatinase—were detected; whereas α-haemolytic activity, proteinase and caseinase were each detected in one isolate only (see Table 6).
Table 3: Antibiotic Resistance Profile of E. coli Strains Isolated from Leafy Vegetables
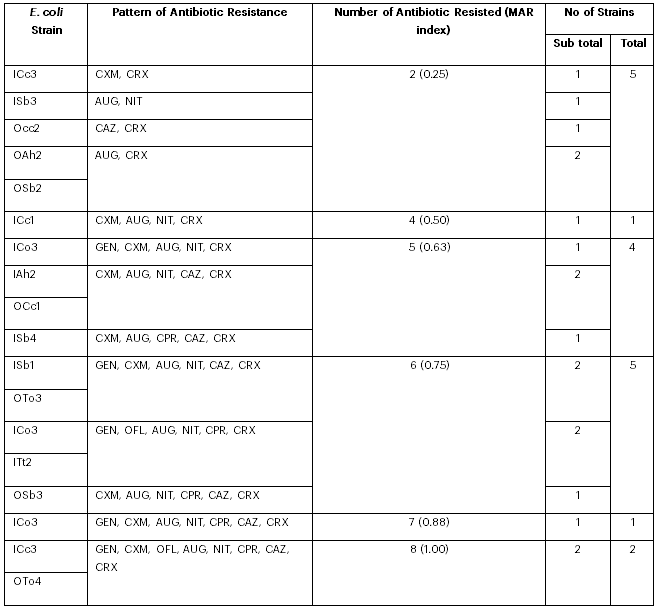
Table 4: Occurrence of Antibiotic Resistance Genes in E. coli Isolates

Key: “+” indicates presence and “–” indicates absence of the gene.
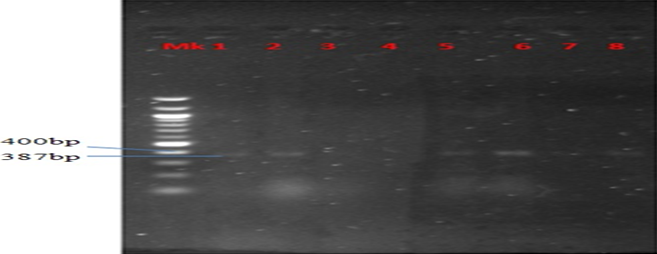
Plate 1: Presence of ParC gene in MAR E. coli strains from leafy vegetables

Plate 2:Presence of Aad2 gene in MAR E. coli strains from leafy vegetables
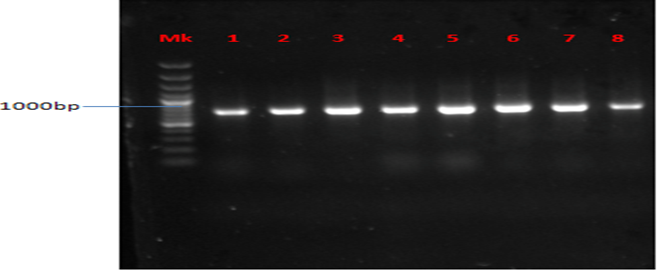
Plate 3: Presence of TEM gene in MAR E. coli strains from leafy vegetables
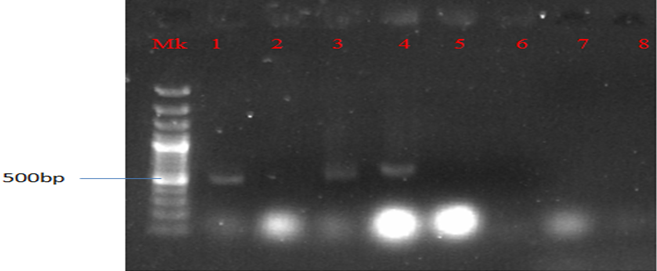
Plate 4: Presence of QnrA gene in MAR E. coli strains from leafy vegetables

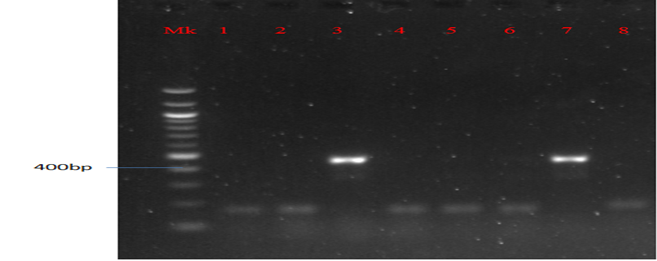
Plate 5: Presence of aac(3)-IV gene in MAR E. coli strains from leafy vegetables
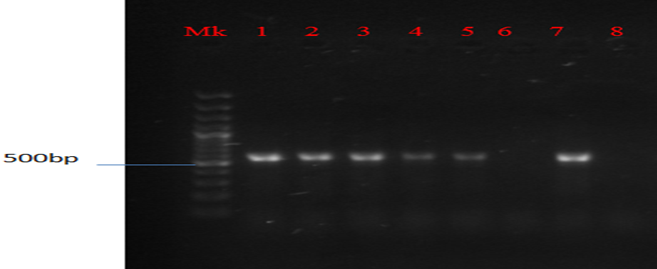
Plate 6: Presence of Ctx-m gene in MAR E. coli strains from leafy vegetables

Plate 7: Presence of shv gene in MAR E. coli strains from leafy vegetables

Plate 8: Presence of ant(2″)-IV gene in MAR E. coli strains from leafy vegetables

Table 5: Occurrence of Virulence Genes in E. coli Isolates

Key: “+” indicates presence and “–” indicates absence of the gene.
Table 6: Enzymatic Activities of MAR E. coli Strains from Leafy Vegetables
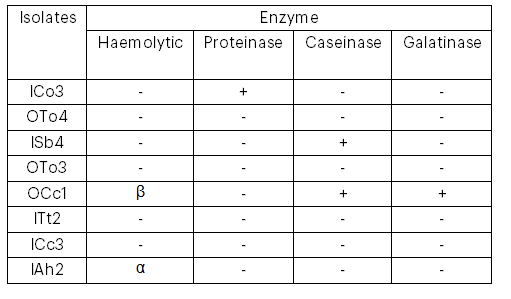
Key: α = alpha haemolysis; β = beta haemolysis; “+” indicates presence; “–” indicates absence.
4. Discussion
The presence of pathogenic microorganisms on vegetables, particularly pathogenic strains of E. coli, is a major public health concern. Previous studies have reported the occurrence of E. coli on vegetable samples (CDR, 1997; Buck & Walcott, 2003; Benyassine et al., 2007), which is attributed to the organism’s ability to survive and proliferate on various vegetable surfaces (Abadias et al., 2012).
In the present study, each isolate demonstrated resistance to at least two antibiotics, confirming that all were multiple antibiotic-resistant (MAR) strains. Moreover, each MAR strain carried at least one of the three virulence genes assayed. The observed MAR patterns among isolates from different vegetables raise serious health concerns. Such patterns may arise from the unrestrained and indiscriminate use of antibiotics in animal treatment and the inclusion of antibiotics in animal feed.
Among the antibiotic resistance genes, the TEM gene was the most prevalent (100% occurrence) and no isolate was free of AR genes. The fimH gene, which encodes type I fimbriae, was present in the majority (62.5%) of the isolates, a finding that is consistent with the work of Paniagua-Contreras et al. (2017), although at a lower frequency than reported by Malekzadegan et al. (2018). Dale and Woodford (2015) have noted that the presence of multiple bacterial virulence factors can influence the extent and severity of infection. Furthermore, Gao et al. (2017) suggested an association between the fimH virulence gene and cystitis cases.
The detection of the usp virulence gene in several isolates is indicative of uropathogenic E. coli, which aligns with the findings of Sepehri et al. (2011), who reported a significant positive association between usp sequences and E. coli strains isolated from patients with inflammatory bowel diseases, such as ulcerative colitis. Shiga toxin (stx1), a major virulence factor characteristic of enterohaemorrhagic E. coli (EHEC), was also detected in all the isolates. Although the stx1 detected in this study is considered less virulent than stx2 (which is more commonly associated with severe human disease; Beutin et al., 2004; Caprioli et al., 2005), its presence remains a cause for concern. This finding is in agreement with Cheun et al. (2010), who reported the presence of Shiga toxin in pathogenic E. coli isolates from patients with diarrhoeal disease in Korea.
A co-association between antibiotic resistance genes and virulence genes was evident. In particular, one isolate that contained all the antibiotic resistance genes screened also harboured all the virulence factors assayed. Notably, AR genes such as ctx-m and shv (markers for β-lactam resistance), together with the virulence factors usp and stx1, were co-present in four isolates. Previous studies have reported significant associations between stx1 and bla_KPC, aac(3) and bla_TEM, and aat with bla_VIM (Pakbin et al., 2021; Koutsoumanis et al., 2020); however, in the current study only one stx1-positive isolate carried bla_KPC, suggesting that such associations may not be universal. The co-presence of virulence and resistance genes likely enhances the survival and persistence of these pathogenic strains in both the host and the environment. Similar co-existence of stx1 and bla_CTX has been reported among E. coli isolates from France and Japan (Valat et al., 2012; Ishii et al., 2005).
Enzymatic profiling of the isolates revealed varied haemolytic and proteolytic activities. For instance, one isolate (designated EOA) exhibited β-haemolysis, which is associated with cell-bound haemolysin production, whereas another isolate (SIB) demonstrated α-haemolysis, indicative of cell-free haemolysin production. Additionally, some isolates exhibited caseinase and gelatinase activities. The presence of these enzymatic activities is a further indication of the pathogenic potential of these isolates; for example, proteases produced by E. coli have been reported to have direct effects on the central nervous system. These findings are consistent with those of Abha et al. (2014), who reported that out of 80 vegetable samples processed, 43 (53.75%) yielded E. coli, with Enterobacter cloacae also being identified as a predominant organism in fresh vegetables
5. Conclusion
The detection of various E. coli strains and the variability in virulence factors among isolates from vegetables represent a significant public health risk, indicative of poor hygienic handling practices. The association of specific virulence genes and enzymatic virulence factors with certain E. coli categories complicates the characterisation of these pathogens. A comprehensive system for assessing the virulence patterns of E. coli isolates—one that examines the full spectrum of virulence genes and corresponding virulence factors—would enhance the characterisation of clinical isolates as well as strains from fresh leafy vegetables, water and environmental sources. Such an approach may provide clearer insights into their pathogenic potential and facilitate the early detection of emerging pathogenic clones. Ultimately, detailed characterisation of E. coli strains based on both their resistance and virulence profiles will inform more effective therapeutic interventions.
Recommendation
Proper cooking or processing of vegetables should be strongly encouraged, as thermal treatment can destroy E. coli. In addition, food should be refrigerated promptly at an appropriate temperature to slow bacterial growth and prevent food poisoning. It is advisable that food be thawed in the refrigerator, under cold running water, or in a microwave immediately prior to cooking. The use of combination therapy may also help reduce the impact of antibiotic resistance. Moreover, consumer education regarding the potential risks associated with consuming fresh vegetables is essential.
Funding
Nil
Conflict of Interest
No competing or conflict of interest to declare.
References
Abadias, M., Usall, J., Anguera, M., Solsona, C., & Vinas, I. (2012). Microbiological quality of fresh, minimally processed fruit and vegetables, and sprouts from retail establishments. International Journal of Food Microbiology, 123(1–2), 121–129.
Abha, D., Namita, J., Rajesh, K. J., & Ahilesh, K. (2014). Molecular characterisation of Escherichia coli isolated from raw vegetable. Advances in Animal and Veterinary Sciences, 2(1), 42–45.
Benyassine, K., Jaa, A., Yahayan, K., & Moulay, M. E. (2007). A simple and rapid detection by PCR of enteropathogenic Escherichia coli in naturally contaminated vegetables. Journal of Rapid Methods in Microbiology, 16, 113–121.
Beuchat, L. R. (2008). Surface decontamination of fruits and vegetables eaten raw: A review (WHO/FSF/98.2). Geneva, Switzerland: World Health Organization.
Beutin, L., Krause, G., Zimmermann, S., Kaulfuss, S., & Gleier, K. (2004). Characterisation of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. Journal of Clinical Microbiology, 42, 1099–1108.
Buck, J. W., & Walcott, R. R. (2003). Recent trends in microbiological safety of fruits and vegetables. Plant Health Progress, 10, 1092–1098.
Caprioli, A., Morabito, S., Brugere, H., & Oswald, E. (2005). Enterohaemorrhagic Escherichia coli: Emerging issues on virulence and modes of transmission. Veterinary Research, 36, 289–311.
CDR. (1997). Hospital outbreak of Escherichia coli O157:H7 associated with a rare phage type, Ontario. Canada Communicable Disease Report, 23, 04–05.
Centers for Disease Control and Prevention. (2014). Outbreaks involving Escherichia coli. Retrieved November 11, 2024, from http://www.cdc.gov/ecoli /outbreaks.html
Centers for Disease Control and Prevention. (2020). E. coli outbreak linked to unknown source 3. Retrieved November 11, 2024, from https://archive.cdc.gov /www_cdc_gov/ecoli/2020/o157h7-11-20/index.html
Cheun, H. I., Cho, S. H., Lee, J. H., Lim, Y. Y., Jeon, J. H., Yu, J. R., Kim, T. S., Lee, W. J., Cho, S. H., Lee, D. Y., Park, M. S., Jeong, H. S., Chen, D. S., Ji, Y. M., & Kwon, M. H. (2010). Infection status of hospitalised diarrhoeal patients with gastrointestinal protozoa, bacteria, and viruses in the Republic of Korea. Korean Journal of Parasitology, 48(2), 113–120.
Dale, A. P., & Woodford, N. (2015). Extra-intestinal pathogenic Escherichia coli (ExPEC): Disease, carriage and clones. Journal of Infection, 71, 615–626.
Dias, J. S. (2012). Nutritional quality and health benefits of vegetables: A review. Food and Nutrition Sciences, 3, 1354–1374. https://doi.org/10.4236 /fns.2012.32167
Eniola, K. I. T., David, O. M., Ajayi, P. O., & Ayo, E. O. (2021). Antibiotic resistance among Escherichia coli from leafy vegetables sold at two markets around Joseph Ayo Babalola University, Ikeji Arakeji, Osun State, Nigeria. Nigerian Journal of Microbiology, 35(2), 5671–5678.
Gao, Q., Zhang, D., Ye, Z., Zhu, X., Yang, W., Dong, L., Gao, S., & Liu, X. (2017). Virulence traits and pathogenicity of uropathogenic Escherichia coli isolates with common and uncommon O serotypes. Microbial Pathogenesis, 104, 217–224.
Grant, J., Wendelboe, A. M., Wendel, A., Jepson, B., Torres, P., Smelser, C., & Rolfs, R. T. (2008). Spinach-associated Escherichia coli O157:H7 outbreak, Utah and New Mexico. Emerging Infectious Diseases, 14, 1633–1636.
Gupta, S. K., Keck, J., Ram, P. K., Crump, J. A., Miller, M. A., & Mintz, E. D. (2008). Part III: Analysis of data gaps pertaining to enterotoxigenic Escherichia coli infections in low and medium human development index countries, 1984–2005. Epidemiology and Infection, 136, 721–738.
Gupta, H., & Malik, R. K. (2007). Incidence of virulence in bacteriocin producing enterococcal isolates. Lait Dairy Science and Technology, 87, 587–601.
Haddadi, K., Moussaoui, F., Hebia, I., Laurent, F., & Le Roux, Y. (2005). Escherichia coli proteolytic activity in milk and casein breakdown. Reproduction, Nutrition, Development, 45(4), 485–496.
Ishii, Y., Kimura, S., Alba, J., Shiroto, K., Otsuka, M., Hashizume, N., Tamura, K., & Yamaguchi, K. (2005). Extended-spectrum beta-lactamase-producing Shiga toxin gene (Stx1)-positive Escherichia coli O26:H11: A new concern. Journal of Clinical Microbiology, 43, 1072–1075.
Johnning, A., Kristiansson, E., Fick, J., Weijdegård, B., & Larsson, D. G. J. (2015). Resistance mutations in gyrA and parC are common in Escherichia communities of both fluoroquinolone-polluted and uncontaminated aquatic environments. Frontiers in Microbiology, 6, Article 3389.
Koutsoumanis, K., Allende, A., Alvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., Davies, R., De Cesare, A., Herman, L., Hilbert, F., Lindqvist, R., Nauta, M., Peixe, L., Ru, G., Simmons, M., Skandamis, P., Suffredini, E., Alter, T., Crotta, M., Ellis-Iversen, J., Hempen, M., Messens, W., & Chemaly, M. (2020). Update and review of control options for Campylobacter in broilers at primary production. EFSA Journal, 18(4), e06090.
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of food. Applied and Environmental Microbiology, 46(1), 165–170.
Lim, P. O., Kim, H. J., & Nam, H. G. (2007). Leaf senescence. Annual Review of Plant Biology, 58, 115–136.
Makhado, U. G., Foka, F. E. T., Tchatchouang, C.-D. K., Ateba, C. N., & Manganyi, M. C. (2022). Detection of virulence gene of Shiga toxin-producing Escherichia coli (STEC) strains from animals with diarrhoea and water samples in the North-West Province, South Africa. Gene Reports, 27, Article 101617.
Malekzadegan, Y., Khashei, R., Ebrahim-Saraie, H. S., & Jahanabadi, Z. (2018). Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infectious Diseases, 18(1), 572.
Natvig, E. E., Ingham, S. C., Ingham, B. H., Cooperband, L. R., & Roper, T. R. (2002). Salmonella enterica serovar Typhimurium and Escherichia coli contamination of root and leaf vegetables grown in soil with incorporated bovine manure. Applied and Environmental Microbiology, 68, 2737–2744.
Olaimat, N. A., & Holley, R. A. (2012). Factors influencing the microbial safety of fresh produce: A review. Food Microbiology, 32(1), 1–19.
Paniagua-Contreras, G. L., Monroy-Perez, E., Rodriguez-Moctezuma, J. R., Dominguez-Trejo, P., & Vaca-Paniagua, F. (2017). Virulence factors, antibiotic resistance phenotypes and O-serogroups of Escherichia coli strains isolated from community-acquired urinary tract infection patients in Mexico. Journal of Microbiology, Immunology and Infection, 50(4), 478–485.
Pakbin, B., Bruck, W. M., & Rossen, J. W. A. (2021). Virulence factors of enteric pathogenic Escherichia coli: A review. International Journal of Molecular Sciences, 22(18), 9922.
Rai, P. K., & Tripathi, B. D. (2007). Microbial contamination in vegetables due to irrigation with partially treated municipal wastewater in a tropical city. International Journal of Environmental Health Research, 17, 389–395.
Rasheed, M. U., Thajuddin, N., Ahamed, P., & Teklemariam, Z. (2014). Antimicrobial drug resistance in strains of Escherichia coli isolated from food sources. Revista do Instituto de Medicina Tropical de São Paulo, 56, 341–346.
Ratchtrachenchai, O. A., Subpasu, S., Hayashi, H., & Ba-Thein, W. (2004). Prevalence of childhood diarrhoea-associated Escherichia coli in Thailand. Journal of Medical Microbiology, 53, 237–243.
Sepehri, S., Khafipour, E., & Bernstein, C. N. (2011). Characterisation of Escherichia coli isolated from gut biopsies of newly diagnosed patients with inflammatory bowel disease. Inflammatory Bowel Diseases, 17, 1451–1463.
Swedan, S., & Alrub, H. A. (2019). Antimicrobial resistance, virulence factors, and pathotypes of Escherichia coli isolated from drinking water sources in Jordan. Pathogens, 8(2), Article 3390. https://doi.org/10.3390/pathogens80203390
Tango, C. N., Choi, N. J., Chung, M. S., & Oh, D. H. (2014). Bacteriological quality of vegetables from organic and conventional production in different areas of Korea. Journal of Food Protection, 77, 411–417.
United Nations Children’s Fund. (2009). Battling diarrhoea. Retrieved November 10, 2024, from https://www.unicef.org/innovation/stories/battling-diarrhoea.html
United Nations Children’s Fund. (2024). Diarrhoea UNICEF data. Retrieved November 12, 2024, from https://data.unicef.org/topic/child-health/diarrhoeal-disease.html
Valat, C., Haenni, M., Saras, E., Auvray, F., Forest, K., Oswald, E., & Madec, J. Y. (2012). CTX-M-15 extended-spectrum beta-lactamase in a Shiga toxin-producing Escherichia coli isolate of serotype O111:H8. Applied and Environmental Microbiology, 78, 1308–1309.
Van Haute, S., Uyttendaele, M., & Sampers, I. (2015). Coagulation of turbidity and organic matter from leafy-vegetable wash-water using chitosan to improve water disinfectant stability. LWT – Food Science and Technology, 64(1), 337–343.
About this Article
Cite this Article
APA
Eniola K.I.T., David O.M., Ajayi P.O., Ajayi O.O. & Adeyemo-Eleyode V.O. (2025). Co-Presence of Antibiotic Resistance Genes and Virulence Genes in Multiple Antibiotic-Resistant Escherichia coli Strains from Leafy Vegetables. In Akinyele B.J., Kayode R. & Akinsemolu A.A. (Eds.), Microbes, Mentorship, and Beyond: A Festschrift in Honour of Professor F.A. Akinyosoye. SustainE
Chicago
Eniola K.I.T., David O.M., Ajayi P.O., Ajayi O.O. and Adeyemo-Eleyode V.O. 2025. “Co-Presence of Antibiotic Resistance Genes and Virulence Genes in Multiple Antibiotic-Resistant Escherichia coli Strains from Leafy Vegetables.” In Microbes, Mentorship, and Beyond: A Festschrift in Honour of Professor F.A. Akinyosoye, edited by Akinyele B.J., Kayode R. and Akinsemolu A.A., SustainE.
Received
10 October 2024
Accepted
7 January 2025
Published
4 February 2025
Corresponding Author Email: kennyeniola@gmail.com
Disclaimer: The opinions and statements expressed in this article are the authors’ sole responsibility and do not necessarily reflect the viewpoints of their affiliated organizations, the publisher, the hosted journal, the editors, or the reviewers. Furthermore, any product evaluated in this article or claims made by its manufacturer are not guaranteed or endorsed by the publisher.
Distributed under Creative Commons CC-BY 4.0
Share this article
Use the buttons below to share the article on desired platforms.














